Abstract
Approximately 8 million people worldwide including 2 million people in the United States are affected by Type 1 Diabetes (T1D) which primarily develops in children under the age of 18. This project aimed to identify shared clinically genetic risk variants and molecular pathways among T1D and other autoimmune diseases such as Rheumatoid Arthritis (RA) , Hashimoto’s Hypothyroidism Disease, and Celiac Disease. The systematic in-depth analysis of GWAS (Genome wide association studies) and dbSNP (single nucleotide polymorphism database) led to the identification of potentially pathogenic coding variants in genes such as PTPN22, FUT2, TYK2 and NCF2. In addition, several non-coding variants were found in regulatory regions of CTLA4, SH2B3, and LRRK2. The structural and functional effect of coding variant of TYK2 was further investigated using the SWISS-MODEL protein modelling platfoirm.These finding contribute to a comprehensive understanding of the genotype-phenotype relationship underlying these common clinically risk variants in pathogenesis of autoimmune diseases and provide a basis for future in vitro and in vivo functional studies using CRISPR (clustered regularly interspaced short palindromic repeats) genome editing strategies.
Introduction
Type 1 Diabetes (T1D) or juvenile diabetes is a chronic disease, characterized by the destruction of pancreatic beta cells, resulting in lifelong loss of insulin secretion. This is a life threatening condition if left untreated and typically managed through continuous monitoring and external insulin administration. T1D often begins with genetic predisposition, resulting in individuals with certain genetic risk variants at higher likelihood of developing the disease. Diagnosis usually occurs at Stage 1 or Stage 2 when two or more T1D associated auto-antibodies are detected in the blood. However, these stages are often late, after the immune system has already started attacking pancreatic beta cells, leading to abnormal blood glucose levels.
There are ongoing efforts to screen for genetic risk factors early that lead to T1D, allowing for preventative measures to be implemented sooner. Previous studies have1,2,3,4 indicated that T1D frequently have co-occurring autoimmune disorders (AIDs) such as Rheumatoid Arthritis (RA) , Hashimoto’s thyroiditis, and celiac disease
The reason co-occurring autoimmune diseases are so prevalent remains unclear, though genetics plays a significant role. This study aims to explore the genetic relationships among T1D and other AIDs to support the development of effective prevention strategies, early screening, intervention and treatment.
Previous studies using immunochip analysis have identified shared genetic risk variants between T1D and RA that include CTLA4, STAT4, IL-2RA5 and primarily focused on HLA associations. However, none of the previous studies has systematically investigated cross-disease and molecular pathways analysis of shared genetic risk variants among T1D, RA, celiac and Hashimoto’s disease. This is the first study to examine non-HLA risk association among four different autoimmune diseases using comprehensive genomic and functional pathway analysis. The goal is to identify shared clinically risk variants or cellular pathways linking Type 1 Diabetes (T1D) with these commonly associated autoimmune diseases.
Genome Wide Association Studies (GWAS) detects association between single nucleotide polymorphism (SNPs) and phenotypic traits using genotypic data collected from microarray or next generation sequencing of whole genome or whole exome sequencing6.
GWAS has successfully identified risk variants or risk alleles for complex diseases such as Type 2 diabetes, Parkinson’s disease, Crohn’s disease, heart diseases and various cancers. It identifies significant association with p value less than 5X10-8, but does not identify causality leaving the biological significance of many variants unknown. The dbSNP database catalogues all human variants including synonyms and non-synonymous variants, insertions and deletions7. It includes both pathogenic (clinically relevant) mutations and common naturally occurring benign mutations. dbSNP database is instrumental in annotating GWAS variants database and filtering out benign variations. All the tools used in this study are open sourced and readily accessible.
This study will help to calculate polygenic risk score which will help in early screening of Type 1 diabetes patients at risk of developing additional autoimmune diseases. The goal of this investigation is eventually to pave the way to potential therapeutic targets for drug discovery research.
Methods
GWAS8 genomic variants datasets for (T1D, RA, Hashimoto’s disease, Celiac disease) were downloaded. Overlap analysis between rsIDs and genes was performed to identify common genetic variants and affected genes amongst all 4 diseases using Venny software tool9. The rsID number is a unique label (“rs” followed by a number) are used by researchers and databases to identify a specific SNP (Single Nucleotide Polymorphism). Gene enrichment analysis was performed for SNPs using Shiny Go10 to identify pathway enrichment for each of the disease. Overlap genes were further analyzed for protein-protein interactions using String database analysis11. String database contains information about protein-protein interactions reported through co-occurence of genes, co-expressions or experimental data. Python programming using Jupyter notebook was used to shortlist clinically relevant variants from a huge dbSNP database12 of 1.5 million variants. rsIDs were input into this script and clinically relevant SNPs were derived with genomic location as output. VaradB13 and G-profiler14 analysis was performed to further annotate non-coding and coding variants common amongst all 4 diseases. Structure modeling for WT and mutant TYK2 was performed using SWISS Model and superimposed using Pymol15.
Methodology is explained in Figure 1.

Results and Discussion
Genomic analysis identified genetic risk alleles and associated genes common among all 4 autoimmune diseases
Genome-wide association studies (GWAS) datasets for all 4 diseases- Type 1 Diabetes, Rheumatoid Arthritis (RA) , Hashimoto’s Hypothyroidism Disease, and Celiac Disease were obtained (Supplementary Data 1). The number of statistically significant risk allelle reported identified by rsIDs and mapped genes were about 710 for T1D, 253 for Celiac, 2231 for Rheumatoid Arthritis and 33 for Hashimoto’s disease. To explore potential shared genetic susceptibility, an overlap analysis of these risk alleles (rsIDs) was conducted using the venny bioinformatics tool. Results are shown as Figure 2A. Two common risk alleles rs72928038-A (BACH2) and rs3184504-C (SH2B3) were found overlapping among all 4 diseases and 48 common alleles were found for T1D, Rheumatoid and celiac diseases. As Hashimoto’s disease dataset was limited and there were only 2 common alleles identified across all 4 diseases, gene mapped with these risk alleles were further extracted from GWAS database tables (Supplementary Data 1) and Venny overlap analysis was performed to analyze common genes across all 4 diseases (Figure 2B and Table 1). Overlapping genes can suggest potential novel biomarkers and drug discovery targets for T1D, Celiac, Hashimoto’s’ disease and RA, however need to be further validated experimentally. Even though these are statistically significant genes and risk alleles, there is a possibility that these risk alleles are present in other autoimmune diseases and other unrelated diseases which cannot be ruled out and out of scope for this work.

*Hashimoto’s disease has a limited dataset in GWAS.
| Gene name | Gene description | Pathway association(s) |
| BACH2 | BTB domain and CNC homolog 2 | B-cell and T-cell differentiation |
| STAT4 | Signal transducer and activator of transcription 4 | Cytokine signaling |
| RNU6-474P | RNA, U6 small nuclear 474, pseudogene | Cytokine signaling |
| CTLA4 | Cytotoxic T-lymphocyte associated protein 4 | Allograft rejection |
| HLA-DQA1 | HLA-DQA1 antisense RNA 1 (MHC Class II molecule) | Antigen presentation to T cells |
| ICOS | Inducible T cell costimulator | T cell activation |
| ATXN2 | Ataxin 2 | RNA processing and translation |
| SH2B3 | SH2B adaptor protein 3 | Cytokine signaling |
| IL2RA | Interleukin 2 receptor subunit alpha | Allograft rejection, cytokine signaling |
Pathway enrichment and gene network interactions analysis indicate JAK-STAT and immune signaling pathway as commonly affected
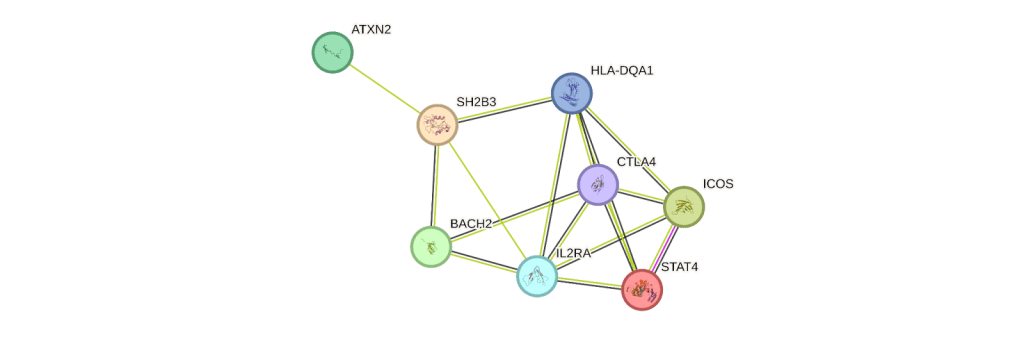
In order to understand the functional significance of these nine overlapping genes, STRING analysis was performed. String analysis extracts functional proteomic analysis for each of these genes and thread these in the protein interaction network (Figure 3). Most of these genes were involved in T cell regulation for inflammation response and immunosuppression. CTLA4 is majorly expressed in T cells and works as a critical regulator in inflammatory response. Patients with RA and T1D are reported to have lower levels of CTLA4 mRNA and protein levels16. IL2RA is expressed high in regulatory T cells and involved in T cell immunosuppression for auto, allo antigens17. These common genes can be novel biomarkers and used to develop polygenic risk score for early detection and prevention of these autoimmune diseases.
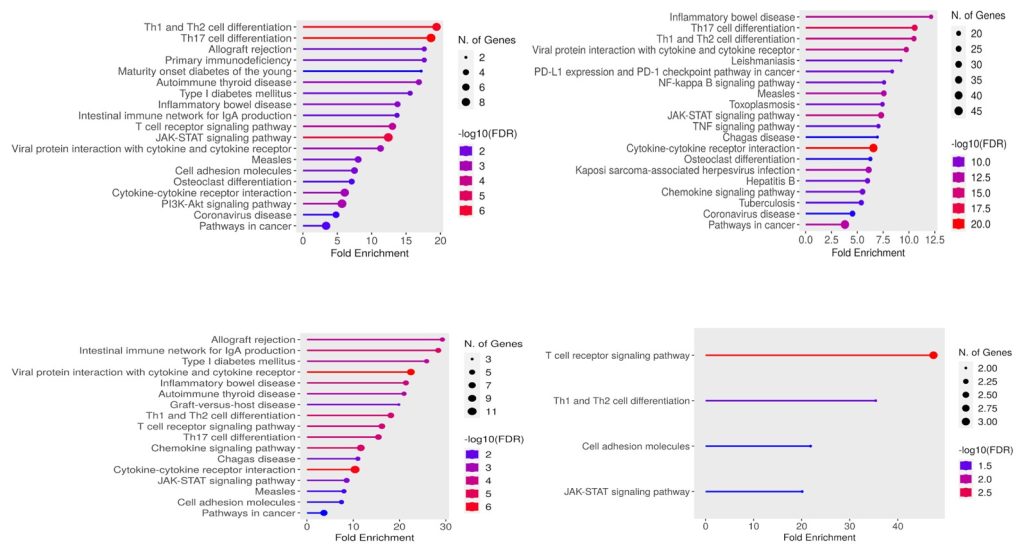
Pathway enrichment analysis for risk allele associated genes for each of the 4 autoimmune diseases revealed several commonly affected pathways. These shared pathways include allograft rejection, T cell differentiation and the JAK-STAT pathway indicating a convergence of immune-related molecular mechanisms across diseases. Corresponding KEGG pathway analysis are shown for T1D (Figure 4A), Rheumatoid (Figure 4B), Celiac (Figure 4C) and Hashimoto’s (Figure 4D). The size of the dots represents the number of genes, and the color represents the significance of the FDR value. FDR is False discovery rate and the higher the value of -log(FDR), smaller is FDR.
Identification and annotation of clinically relevant genetic risk alleles
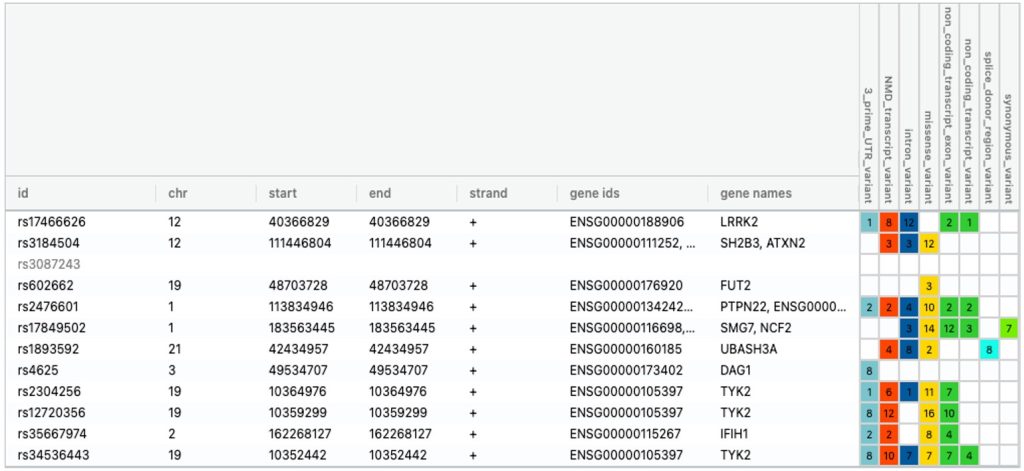
To address the challenge of analyzing a large dbSNP data containing over 1.5 million SNPs (single nucleotide polymorphism), a custom python program was developed (Supplementary method) to systematically identify clinically significant genetic risk variants associated with 4 autoimmune diseases. Statistically significant risk alelles (rsIDs) derived from GWAS dataset (Supplementary Data 1) were cross-referenced with dbSNP database to retain only those annotated with clinical significance (Supplementary Data 2). All other rsIDs lacking clinical annotations in dbSNP were filtered out. This stringent filtering resulted in a reduced and clinically significant SNP dataset consisting of 21 variants for T1D, 69 variants for RA, 11 variants for Celiac and 2 variants for Hashimoto’s. Venny analysis was performed again to analyze common variants among all 4 autoimmune diseases and are all listed in Figure 5.
Chromosomal genomic location and annotation were also extracted for each clinically significant common risk alleles.. The gene names, genomic coordinates along with annotations for each of these genetic risk variants in all 4 diseases are shown in Figure 5. Further, VaradB analysis was used to validate these common clinically significant risk variants. VARAdb is a useful resource for selecting potential functional variations and interpreting their effects on human diseases and biological processes.
This helped in identifying coding variant (gene: TYK2) and non-coding clinically relevant variants (3’UTR in CTLA4 and enhancer in SH2B3) common amongst all 4 diseases.
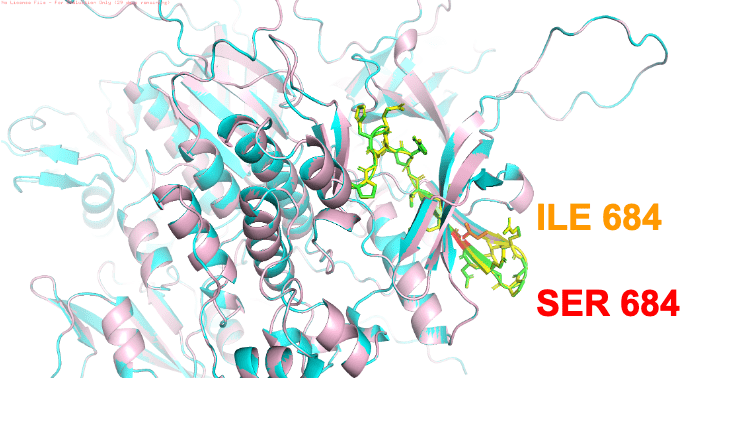
Clinically relevant mutation identified for TYK2 potentially alters protein functionTyrosine Kinase 2 (TYK2) variant rs12720356 was identified as the most frequently associated clinically significant risk genetic allele amongst all 4 autoimmune diseases. Tyrosine kinase 2 (TYK2) is an intracellular enzyme that mediates immune signaling and inflammatory signaling pathways18. This coding variant leads to ILE684 to SER684 mutation. Wild type TYK2 and mutant TYK2 ILE684 to SER684 were modeled using SWISS model and aligned using pymol (cyan ribbon diagram: wt TYK2 (4PO6 with 1.99Å resolution), pink ribbon diagram : mutant TYK2. RMSD of two overlapped structures was very low and structures aligned pretty well indicating this mutation is not likely to perturb protein structure. This mutation probably affects function and is earlier reported to be catalytically inactive and has impaired signaling using cell based in vitro kinase activity assay19 . TYK2 activates a series of transcription factors called signal transducer and activator of transcription (STAT).8 Activated STATs promote expression of cytokines and cellular processes such as cellular division, differentiation and death.8 By binding to specific receptors, cytokines signal through TYK2 to regulate the immune system. These cytokines include IL-12, IL-23 and Type I IFNs, which are critical in driving the function of Th1 cells, Th17 cells and the innate immune response20. Activation by type I IFNs signaling through TYK2 in the B cell impacts pathways important in autoimmunity such as B cell differentiation, antibody production, and immunoglobulin isotype class switching.
Summary and Conclusions
This study analyzed GWAS data across four autoimmune diseases – Type 1 Diabetes, Rheumatoid Arthritis (RA), Hashimoto’s thyroiditis, and celiac disease to identify overlapping genetic variants and molecular pathways involved in disease susceptibility. A total of 3,227 significant risk variants were examined and comparative analysis revealed two shared variants across all 4 diseases (rs72928038-A in BACH2 and rs3184504-C in SH2B3) with 48 additional alleles common among T1D, Celiac and RA.
Further, gene overlap analysis identified nine common genes, many of which are involved in T-cell regulation, inflammatory response, and immunosuppression. Pathway enrichment and network analysis highlighted the JAK-STAT signalling and immune response pathway as commonly dysregulated, emphasizing their critical role in autoimmunity.
To prioritize clinically relevant risk variants, a filtering pipeline using dbSNP and varadB was implemented. This yielded variants in coding regions for genes such as PTPN22, FUT2, TYK2 and NCF2 as well as non-coding variants in regulatory regions of CTLA4, SH2B3, and LRRK2. TYK2 ILE684 to SER684 mutation was identified as common among all 4 diseases and can be a potential target for drug discovery. Structural modeling suggested this mutation does not alter protein structure significantly but likely impairs TYK2 function, consistent with its previously reported catalytic inactivity.
Future directions
The potential limitation of using GWAS dataset is biased associations as 79% of the population in this study are of European origin. However, it will be good to explore other databases such as OMIM and HGMD in future as these are not very comprehensive yet for autoimmune diseases.It is imperative to validate these findings using functional experimental studies in cell lines or animal models to understand the mechanism of these variants in regulating immune signaling and disease pathogenesis. This clinically relevant genetic risk alleles gene set can be used to develop polygenic risk scores for early diagnosis of celiac, rheumatoid arthritis and Hashimoto’s disease in T1D patients and help in development of personalized treatment plans.
References
- Frommer, L., & Kahaly, G. J. (2020). Type 1 diabetes and associated autoimmune diseases. World J Diabetes, 11(11), 527–539. https://doi.org/10.4239/wjd.v11.i11.527 [↩]
- Marquez, A., & Martin, J. (2022). Genetic overlap between type 1 diabetes and other autoimmune diseases. Semin Immunopathol, 44(1), 81–97. https://doi.org/10.1007/s00281-021-00885-6 [↩]
- Popoviciu, M. S., Kaka, N., Sethi, Y., Patel, N., Chopra, H., & Cavalu, S. (2023). Type 1 Diabetes Mellitus and Autoimmune Diseases: A Critical Review of the Association and the Application of Personalized Medicine. J Pers Med, 13(3). https://doi.org/10.3390/jpm13030422 [↩]
- Strakova, V., Elblova, L., Johnson, M. B., Dusatkova, P., Obermannova, B., Petruzelkova, L., Kolouskova, S., Snajderova, M., Fronkova, E., Svaton, M., Lebl, J., Hattersley, A. T., Sumnik, Z., & Pruhova, S. (2019). Screening of monogenic autoimmune diabetes among children with type 1 diabetes and multiple autoimmune diseases: is it worth doing? J Pediatr Endocrinol Metab, 32(10), 1147–1153. https://doi.org/10.1515/jpem-2019-0261 [↩]
- Marquez, A., Kerick, M., Zhernakova, A., Gutierrez-Achury, J., Chen, W. M., Onengut-Gumuscu, S., Gonzalez-Alvaro, I., Rodriguez-Rodriguez, L., Rios-Fernandez, R., Gonzalez-Gay, M. A., Coeliac Disease Immunochip, C., Rheumatoid Arthritis Consortium International for, I., International Scleroderma, G., Type 1 Diabetes Genetics, C., Mayes, M. D., Raychaudhuri, S., Rich, S. S., Wijmenga, C., & Martin, J. (2018). Meta-analysis of Immunochip data of four autoimmune diseases reveals novel single-disease and cross-phenotype associations. Genome Med, 10(1), 97. https://doi.org/10.1186/s13073-018-0604-8 [↩]
- Cerezo, M., Sollis, E., Ji, Y., Lewis, E., Abid, A., Bircan, K. O., Hall, P., Hayhurst, J., John, S., Mosaku, A., Ramachandran, S., Foreman, A., Ibrahim, A., McLaughlin, J., Pendlington, Z., Stefancsik, R., Lambert, S. A., McMahon, A., Morales, J.,…Harris, L. W. (2025). The NHGRI-EBI GWAS Catalog: standards for reusability, sustainability and diversity. Nucleic Acids Res, 53(D1), D998–D1005. https://doi.org/10.1093/nar/gkae1070 [↩]
- Sherry, S. T., Ward, M. H., Kholodov, M., Baker, J., Phan, L., Smigielski, E. M., & Sirotkin, K. (2001). dbSNP: the NCBI database of genetic variation. Nucleic Acids Res, 29(1), 308–311. https://doi.org/10.1093/nar/29.1.308 [↩]
- https://www.ebi.ac.uk/gwas/ [↩]
- https://bioinfogp.cnb.csic.es/tools/venny/ [↩]
- https://bioinformatics.sdstate.edu/go/ [↩]
- https://string-db.org/ [↩]
- https://www.ncbi.nlm.nih.gov/snp/ [↩]
- http://www.licpathway.net/VARAdb/ [↩]
- https://biit.cs.ut.ee/gprofiler/gost [↩]
- https://www.pymol.org/ [↩]
- Hossen, M. M., Ma, Y., Yin, Z., Xia, Y., Du, J., Huang, J. Y., Huang, J. J., Zou, L., Ye, Z., & Huang, Z. (2023). Current understanding of CTLA-4: from mechanism to autoimmune diseases. Front Immunol, 14, 1198365. https://doi.org/10.3389/fimmu.2023.1198365 [↩]
- Lykhopiy, V., Malviya, V., Humblet-Baron, S., & Schlenner, S. M. (2023). “IL-2 immunotherapy for targeting regulatory T cells in autoimmunity”. Genes Immun, 24(5), 248–262. https://doi.org/10.1038/s41435-023-00221-y [↩]
- Li, Z., Rotival, M., Patin, E., Michel, F., & Pellegrini, S. (2020). Two common disease-associated TYK2 variants impact exon splicing and TYK2 dosage. PLoS One, 15(1), e0225289. https://doi.org/10.1371/journal.pone.0225289 [↩]
- Li, Z., Gakovic, M., Ragimbeau, J., Eloranta, M. L., Ronnblom, L., Michel, F., & Pellegrini, S. (2013). Two rare disease-associated Tyk2 variants are catalytically impaired but signaling competent. J Immunol, 190(5), 2335–2344. https://doi.org/10.4049/jimmunol.1203118 [↩]
- Muromoto, R., Oritani, K., & Matsuda, T. (2022). Current understanding of the role of tyrosine kinase 2 signaling in immune responses. World J Biol Chem, 13(1), 1–14. https://doi.org/10.4331/wjbc.v13.i1.1 [↩]






