Abstract
This review explores genetic polymorphisms in East Asians that influence tuberculosis susceptibility, focusing on Nramp1, HLA-DR2, VDR, and TLR. Genetic polymorphisms can be a great indicator of probability of disease. Mycobacterium tuberculosis(TB), is an airborne disease responsible for 1.5 million deaths per year. It is especially prevalent within East Asian nations. In this study, East Asia includes Southeast Asian nations such as China, Korea, and Japan, as well as Southeast Asian nations such as Malaysia and others. East Asian nations have a disproportionately higher number of TB patients, with Southeast Asia containing 45% of TB cases despite only containing 8.5% of the world’s population. Although various factors could contribute to this susceptibility, one of the most prominent factors is genetic polymorphisms. The reasons for the existence of these polymorphisms within East Asians specifically still remains unclear, but it can be inferred that these polymorphisms are advantageous in some other ways. Several studies have discovered specific polymorphisms within Nramp1, HLA-DR2, VDR, TLR and their impacts on TB susceptibility within East Asians. These polymorphisms often impact the immune system, as it impacts the body’s ability to combat TB. This review explores key polymorphisms and the potential reasons they create susceptibility to TB. Discovering the polymorphisms that cause susceptibility could be crucial to the prevention of TB. Technology could be used to determine the chance of an individual with a particular polymorphism contracting TB, and prevention measures taken. Most studies tend to focus on either a specific region, or a specific polymorphism. There is a lack of research reviewing various polymorphisms while discussing the entirety of East Asia. This review will address this gap in knowledge.
Keywords: Tuberculosis (TB), genetic polymorphisms, Nramp1, HLA-DR2, VDR, TLR, immune system, East Asia, risk factors
1. Introduction
Mycobacterium tuberculosis is a pathogenic bacterium that primarily infects the lungs and causes tuberculosis, a disease responsible for approximately 10 million cases and 1.5 million deaths worldwide each year. Tuberculosis (TB) is most prevalent within low- and middle-income nations, especially within East Asian nations1. Symptoms of TB include a cough that contains mucus sometimes with blood in it, weight loss, fever, a loss of appetite, and others. TB spread could be characterized with body aches in other locations. The risk factors of tuberculosis include HIV, Diabetes, Malnutrition, Poverty, Tobacco, Alcohol, Genetics and others2. Mycobacterium tuberculosis spreads via airborne infections between individuals with TB. When TB enters the body, the innate immune system responds. This consists mostly of macrophages (Fig. 1). The macrophage consumes the TB cell and it undergoes phagocytosis, the process of breaking down the bacteria using enzymes and various chemical reactions. Parts of the bacteria are then attached to the macrophage’s cell membrane to signal antibody production. Adaptive immune system cells, including T-cells and B-cells then create antibodies to fight against the TB bacteria, taking about 4-7 days. Within 90% of TB cases, macrophages and T-cells either eliminate the TB bacterium, or the TB bacterium lies dormant in a state called Latent TB, where TB symptoms are inactive. Under 5% of other instances, TB overpowers macrophages and T-cells and a state called active TB is created, where symptoms of TB start to infect the host. In 5% of cases, Latent TB can be reactivated to create secondary active TB. Latent TB is unremovable from the host in most cases3.
Intriguingly, 45% of TB cases occur in Southeast Asian (SEA) Nations alone, even though SEA is only 8.5% of the world’s population4. This paper focuses on the genetic risk factors within Asians that specifically impacts the innate immune system, discussing specific polymorphisms within genes that lead to tuberculosis within East Asians. Understanding the causes of tuberculosis could lead to enhanced prevention programs targeted to those who have a high probability of contracting tuberculosis.
It is crucial to consider the genetic polymorphisms of tuberculosis as it can act as an indicator for tuberculosis. By finding the polymorphisms that cause tuberculosis, health facilities can adequately identify potential tuberculosis patients and prevent diseases. This research aims to review the question: “To what extent do the prevalence of certain genetic polymorphisms influence the probability of contracting tuberculosis within different regions?”

2. Results
There are many processes within macrophages that play crucial roles in fighting and disrupting the tuberculosis bacterium. Genetic variation in the molecules involved in these processes could therefore create advantageous or disadvantageous situations against tuberculosis. Although there are many variations within different types of genes in these processes that could potentially alter an individual’s susceptibility to tuberculosis, this paper focuses on four main molecules and their associated polymorphisms that may play a significant role in increasing susceptibility to tuberculosis (Fig 1.):The polymorphisms within Nramp1 that will be discussed are the D543N allele, 5’(GT)n as well and the 3’UTR polymorphism. The second molecule is HLA-DR2. The polymorphisms in HLA-DR2 discussed is DRB1 and specifically its serotypes DRB1*1501 and DRB1*1502. The third structure is the vitamin D receptor (VDR). The polymorphism in VDR that will be discussed is FokI. The final structure discussed is the Toll-Like Receptor. The polymorphisms discussed will be LR2 2258 A/G and TLR4 896 A/G. Potential reasons for TB susceptibility are summarized in Table 3.
2.1 Natural Resistance Associated Macrophage Protein 1
Natural Resistance Associated Macrophage Protein 1 (Nramp1) is a protein involved in affecting nutritional immunity within macrophages during phagocytosis. Nutritional immunity is the process of transporting resources away from a foreign substance to limit the invader’s nutrients and resources to allow the macrophage to swiftly destroy foreign invaders during phagocytosis. When foreign substances have access to crucial resources, despite being surrounded by the macrophage, they can struggle and impede phagocytosis. This can therefore cause a slowing of phagocytosis and even the destruction of the macrophage. Nramp1 attempts to prevent this through nutritional immunity. Nramp1 is specifically in charge of transporting Iron (II) and Manganese (II) away from bacteria using active transport5. Therefore, a polymorphism that disrupts Nramp1 function is likely to disrupt macrophage function and increase the risk of tuberculosis. The polymorphisms focused on are the D543N allele, 3’UTR polymorphism, and 5’(GT)n polymorphism.
2.1.1 3’UTR
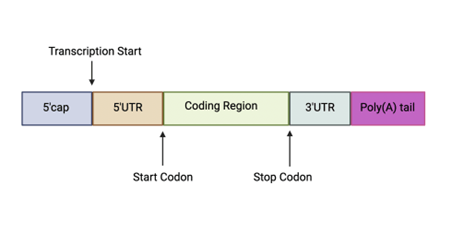
The 3’ untranslated regions (3’UTR) are sections within mRNA that remain untranslated after translation to a protein. Other untranslated regions include 5’ untranslated region, 5’cap, and poly(A)tail. These regions regulate important functions such as translation, mRNA stability, and mRNA localization (Fig. 2). Protein-protein interactions (PPI) mediated by the 3’UTR can allow the genetic information encoded within 3’UTR to be sent to proteins6. This means that a 3’UTR polymorphism could have impacts on Nramp1 function that might allow susceptibility to TB.
Figure 2: RNA processing after introns are removed. 3’UTR, the coding region, 5’cap, 5’UTR, and Poly(A) tail are placed structurally.
2.1.2 D543N
The D543N allele refers to when an aspartic acid at position 543 in the Nramp1 protein is replaced by asparagine. This is known as a missense mutation, which could alter protein function. This alteration could therefore alter Nramp1 in a way that impedes Nramp1’s response to TB.
2.1.3 5’(GT)n
The 5’(GT)n polymorphism refers to when guanine and thymine (GT) occurs, in a repetitive sequence, n times at the 5’ end of a DNA sequence. It is not directly connected to the 5’UTR region. This is known as microsatellite repeats and has the potential to alter the function of Nramp1.
2.1.4 3’UTR, D534N, 5’(GT)n and the polymorphisms associated with TB
In a study of 227 TB patients and 516 controls, individuals with the 3’UTR polymorphism were found to be 35% more likely to have TB compared to those without it (OR = 1.35, 95% CI: 1.17–1.54). D543N and 5’(GT)n polymorphisms are also related to a 25% and a 31% increase in TB susceptibility respectively. These findings suggest that these polymorphisms increase the likelihood of TB, likely by interfering with Nramp1 phagocytic function7. In a meta-analysis conducted with 35 article found that a tuberculosis patient with a D543N polymorphism was 31% more likely to contract tuberculosis than ones without it(OR = 1.31). Similarly, individuals with a 3’UTR polymorphism were 45% more likely to contract tuberculosis than ones without it (OR = 1.45) in East Asia. This consistent association across studies reinforces the role of D543N as a potential genetic risk factor, especially in East Asian populations where the polymorphism appears more prevalent8. A study conducted found that 3’UTR polymorphisms only had a stronger correlation to TB within Asian populations, compared to Africans, South Americans, and Caucasians9. Similarly, neither 3’UTR nor D543N polymorphisms had associations within TB in European populations8. This aligns within the view that Europeans have developed natural resistances to TB compared to Asians10.
Another review of Nramp1 polymorphisms indicated a higher odds ratio for Asians than overall individuals in 3’UTR and 5’(GT)n, but indicated no difference for D543N (Table 1).
Table 1: Nramp1 polymorphisms and their odd ratios of being in tuberculosis patients compared to non-tuberculosis patients11.
| Polymorphisms | Odds Ratio (95% Confidence Interval) | |
| Overall | Asians | |
| 3’UTR | 1.33 (1.08-1.63) | 1.46 (1.10–1.94) |
| D543N | 1.67 (1.36-2.05) | 1.65 (1.29–2.12) |
| 5’(GT)n | 1.32 (1.03-1.68) | 1.86 (1.33–2.62) |
2.2 Human Leukocyte Antigen – DR Isotope Class 2
Human Leukocyte Antigen – DR Isotope Class 2 (HLA-DR2) is an antigen presenting molecule. It presents peptides to CD4 receptors of T-cells for antibody production, specifically encoding MH Class 2 molecules12. Antigen production is crucial in combating diseases because the adaptive immune system is able to produce cells that can specifically respond to TB. A polymorphism within HLA-DR2 could potentially impede the antigen presenting process, impacting whether or not the adaptive immune system is able to properly respond to TB. The polymorphism DRB1*15 and its serotypes (most prominently DRB1*1501 and DRB1*1502) within HLA-DR2 are the most common serotypes within TB patients, however other DR2 polymorphisms also have correlations with TB.
2.2.1 HLA-DRB1 Subtypes
HLA-DRB1 subtypes were most commonly seen in Northeast Asians, Southeast Asians, North Americans, and South Americans, with a consistent frequency of over 20% in many regions. Europe (West, North, and Southeast), Africa(North and Sub-Saharan), and Southwest Asia had significantly lower frequencies of HLA-DRB1*1513. HLA-DRB1*15, a subtype commonly associated with TB, has approximately a 28% frequency in Malaysia14 and over 20% in several East and Southeast Asian populations13. A study conducted with 289 healthy control subjects and 153 TB patients discovered that those with some serotype of HLA-DR2 were more frequently present within tuberculosis patients with 61.82% of those with TB having some type of HLA-DR2, and only 42.86% in control groups without TB15. Another study conducted within Asian Indians found that 90% of pulmonary TB patients had DRB1*1501 allele, while only 65.2% of healthy control subjects had it. Similarly, DRB1*1502 allele had a 64.1% frequency within tuberculoid leprosy patients, while only a 39.1% frequency in control subjects16.
2.3 Vitamin D Receptor
Vitamin D receptor (VDR) is a cytoplasmic receptor that when bound by its ligand activates genes after binding. Vitamin D is prominent within macrophages in activating genes such as cathelicidin antimicrobial peptide (CAMP), a peptide that creates inhibitory effects to bacteria; Interleukin-10, a cytokine that modulates cell homeostasis and inflammation; and many other functions crucial for immune response. There is evidence that those with Vitamin D deficiencies are likely to contract diseases, specifically upper respiratory diseases and autoimmune diseases17. The polymorphism FokI is the main polymorphism impacting TB susceptibility18.
2.3.1 FokI polymorphisms
A FokI polymorphism refers to change in a single nucleotide(C>T) at the start codon (ATG) of the VDR in exon 2. This can result in two forms of protein, the longer or shorter form. The longer form contains 427 amino acids, encoded by the “f allele”. The shorter form is encoded by the “F” allele with 424 amino acids. The shorter form has been found to increase transcriptional activity of the VDR18.
A meta-analysis discovered that there is 32% higher chance of contracting TB within FokI homozygote models (OR = 1.32) and 26% higher in the recessive model compared to non-TB individuals (OR = 1.26) . East Asians individuals with this polymorphism had a slightly higher likelihood of contracting TB19. Similarly, another meta-analysis found that there is a 35% higher likelihood of a TB patient having FokI compared to controls. This study also found that East Asian TB Patients had a 42% higher likelihood of having FokI polymorphisms compared to healthy controls compared to control groups, further suggesting ethnic implications20.
2.4 Toll-like Receptor
Figure 3: The interaction between a ligand and TLR, resulting in the gene activation of type I interferon genes.
Toll-like Receptors (TLRs) are membrane-bound receptors that are often situated in the innate immune system, especially macrophages. Toll-like receptors (TLRs) detect Pathogen-associated molecular patterns (PAMPs), molecules from pathogens like Mycobacterium tuberculosis, and Damage-associated molecular patterns (DAMPs), signals from damaged host cells. These patterns trigger immune responses. Polymorphisms in TLRs may impair this detection, weakening the body’s defense against TB. TLR triggers a cascade inside the immune cell once it binds to its ligand. Transcription factors such as NF-κB are activated, in turn producing pro-inflammatory cytokines and type I interferons. These molecules can activate inflammation to combat pathogens and play crucial roles in the immune system response to pathogens. TLR can also help mature antigen presenting molecules, such as HLA-DR2, therefore supporting the adaptive immune system21. Polymorphisms in TLRs could therefore impact the production of pro-inflammatory cytokines and type I interferons therefore either strengthen or weaken the immune system response to TB. The polymorphisms focused on are TLR2 2258 A/G and TLR4 896 A/G.
2.4.1 TLR2 2258 A/G
TLR2 2258 A/G is an adenine allele at the 2258 position within receptor 2 of TLR that has been replaced by guanine in its genetic sequence.
2.4.2 TLR4 896 A/G
TLR4 896 A/G is an adenine allele at the 896 position within receptor 4 of TLR that has been replaced by a guanine in its genetic sequence.
2.4.3 TLR2 2258 A/G and TLR4 895 A/G associations with TB
There is a 17% higher chance that a TB patient has TLR2 2258 A/G than the chance for a healthy control patient to have TLR2 2258 A/G.13. TLR2 G2258A had an odds ratio of 5.82 for those with TB compared to those without indicating strong association with TB. In an ethnic subgroup analysis, it was found that Asians had increased risk compared to other ethnicities with an odds ratio of 2.95 with a 91% higher likelihood of contracting TB in Asian Indians15’22. This data indicates that TLR2 2258 G/A and TLR4 896 A/G are closely linked with tuberculosis.
Table 2: Structures and their potential contributions to TB susceptibility. Summarizes immune-related structures—such as Nramp1, HLA-DR2, VDR, and Toll-like Receptors (TLRs)—and explains how genetic polymorphisms may impair their functions, weakening host defense against tuberculosis.
| Structures | Potential Contributions to TB Susceptibility |
| Natural Resistance Associated Macrophage Protein 1 (Nramp1) | Involved in nutritional immunity, which is the process of transferring nutrients, specifically FeII and MnII away from a foreign invader during phagocytosis. A polymorphism can disrupt the process allowing the bacteria to maintain more FeII and MnII leading to a slower or failed process of phagocytosis. |
| Human Leukocyte Antigen – DR Isotope Class 2 (HLA-DR2) | Involved in presenting antigens to T-cells. A polymorphism could therefore disrupt antibody production and weaken the adaptive immune system response. |
| Vitamin D Receptor (VDR) | VDR binding activates genes such as cathelicidin antimicrobial peptide (CAMP) and Interleukin-10 ( IL-10) that play crucial roles in immune system response. A polymorphism could therefore disrupt the activation of these genes leading to weaker immune responses. |
| Toll-Like Receptors (TLRs) | Responds to molecular patterns associated with pathogen presence by producing pro-inflammatory cytokines and type I interferons, playing a crucial role in inflammation. A polymorphism in TLRs could therefore disrupt the production of these molecules, weakening immune response. |
3. Discussion
This review presented the polymorphisms Nramp1, HLA-DR2, VDR, TLR and their correlation to TB within East Asia, using multiple studies and meta-analyses. As research surrounding the aforementioned polymorphisms and many other polymorphisms continue, the ability to prevent and combat TB increases. Scientists can create comprehensive systems to discover a patient’s chances of contracting TB, allowing the implementation of prevention measures. As gene editing and other technologies advance, discovering the genetic risk factors for TB could prevent the deaths of many.
Nevertheless, there are various limitations to this discussion. Firstly, it is unclear why these polymorphisms are predominantly present within East Asians. These polymorphisms are likely evolutionary traces that serve other purposes catered specifically towards conditions primarily faced within East Asia. Because of their impact on the immune response to tuberculosis, they were likely developed in order to grant some type of advantage against other pathogens that may be more prevalent within East Asia. Further research is needed to discover the geographic-specific nature of these polymorphisms. If scientists blindly modify these polymorphisms in order to prevent TB, it is likely that other pathogens would impact humans more significantly due to the absence of these polymorphisms. Sociopolitical and environmental factors may also contribute to the high prevalence of TB in East Asia. Urban crowding, poor ventilation, limited healthcare access in rural areas, and inconsistent public health infrastructure can exacerbate TB transmission and progression. East Asian nations could often have significant disparities in health infrastructure and resources access that acts as risk factors for TB. Issues such as childhood wasting could exacerbate disparities in health resource access thus leading to higher TB prevalence23. Consistently, specific genetic polymorphisms such as Nramp1 543N and 3’UTR, HLA-DRB1 1501/1502, and many others are significantly associated with increased TB susceptibility in East Asian population.These studies find that these polymorphism typically create an increase from 25% to 45%, indicating that these polymorphisms impair immune-related processes. Furthermore, differences emerge around the strength and consistency of these associations across different regional groups. For instance, the 3’UTR polymorphisms in Nramp1 shows strong correlations in East Asian populations but not in Europeans or Africans24’9, suggesting that other variables such as urban lifestyle, health disparities, and others could influence whether these polymorphisms lead to TB prevalence. Similarly, while DRB11501 is consistently linked with TB in Asian Indian studies, the extent of its association varies by region and sample size25. Crucially, a gap in research is the lack of studies that aim to explore the mechanisms as to why these polymorphisms influence TB susceptibility. Most studies are correlational in nature. Future research can aim to explore these mechanisms in order to discover additional information on TB. Next, the polymorphisms and genes discussed are only correlational. It is unclear exactly why polymorphisms such as 3’UTR, D543N, FokIand TLR2 2258 A/G lead to susceptibility to TB. Further research is needed to discover the concrete mechanism behind their influence on TB.
Finally, meta-analyses and studies conducted were primarily set within accessible locations and hospitals mostly within urban areas. This could lead to certain less developed nations and areas being excluded from these studies, leading to a less accurate representation of East Asia as a whole. Further research should be conducted within these areas to determine the exact influence of polymorphisms across the whole of East Asia.
4. Methods
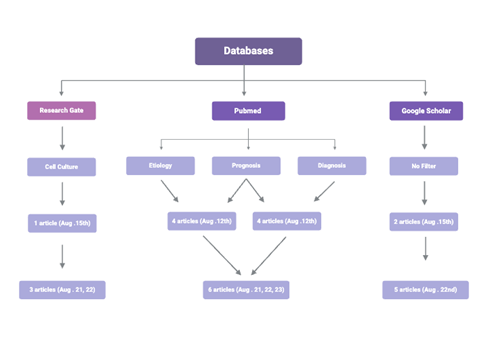
Research articles were obtained from 3 databases: Google Scholar, ResearchGate, and PubMed Clinical Inquiries (Fig. 4). On Aug 12th, PubMed Clinical Inquiries was used with filters ‘prognosis’, ‘diagnosis’, and ‘etiology’ at both a ‘broad’ and ‘narrow’ scope with the search terms ‘genetic susceptibility’ and ‘tuberculosis’. 4 articles were picked based on relevance. On Aug 15th, 2024, research was conducted with Google Scholar using the search terms ‘polymorphisms’, ‘tuberculosis’, and ‘Asia’. Articles were selectively picked based on relevance, resulting in 2 chosen articles. ResearchGate was used with the filter ‘cell culture’ on Aug 15th, 2024, with search terms ‘polymorphisms’, ‘tuberculosis’, and ‘Asia’. Articles were selectively picked based on relevance, and 1 article was chosen. On Aug 15th, the process used on Aug 12th with PubMed Clinical Inquiries was used but instead with the search terms ‘Nramp 1’ and HLA-DR2′. 5 articles were chosen based on relevance. On August 21st, it was decided that the main focus will be NRAMP 1, HLA-DR2, VDR, and TLR gene polymorphisms. This removed 9 articles from the 15 collected. 5 additional articles were found on August 21st, 22nd, and 23rd with PubMed, 2 with ResearchGate, and 5 with Google Scholar using search terms Nramp1, HLA-DR2, VDR, TLR, East Asia, and tuberculosis (Fig. 4).
Figure 4: The research process going from the databases used, to the filters used, to when and how many articles were discovered from each database.
5. Acknowledgements
This was written with the help of professor Dr. Arri Eisen from Emory University and Pano Education. Without their help, this paper would not be possible. Special thanks for my mother and father for raising me and developing in me an interest for biology.
References
- S. Kant, V. Mitta, A. K. Mavi, P. Singh. Tuberculosis: An Overview and Review of Literature. ResearchGate; Springer Nature.https://www.researchgate.net/publication/353637449_Tuberculosis_An_Overview_and_Review_of_Literature. (2021, July 31). [↩]
- P. Narasimhan, J. Wood, C. R. MacIntyre, D. Mathai.Risk Factors for Tuberculosis. Pulmonary Medicine, 2013, 1–11. https://doi.org/10.1155/2013/828939. (2013). [↩]
- E. H. Tobin, & D. Tristram. Tuberculosis. Nih.gov; StatPearls Publishing. https://www.ncbi.nlm.nih.gov/books/NBK441916/.(2024, March 20). [↩]
- V. Bhatia, S. Rijal, M. Sharma, A. Islam, A. Vassall, A. Bhargava, A. Thida, C. Basri, I.i Onozaki, M. Pai., M. Rezwan, N. Arinaminpathy, P. Chandrashekhar, R. Sarin, S. Mandal, Raviglione, M. Ending TB in South-East Asia: flagship priority and response transformation. The Lancet Regional Health – Southeast Asia, 18, 100301–100301. https://doi.org/10.1016/j.lansea.2023.100301. (2023). [↩]
- F. Canonne‐Hergaux, S. Gruenheid, G. Govoni, P. Gros. The Nramp1 Protein and Its Role in Resistance to Infection and Macrophage Function. Proceedings of the Association of American Physicians, 111(4), 283–289. https://doi.org/10.1046/j.1525-1381.1999.99236.x(1999). [↩]
- C. Mayr. What Are 3′ UTRs Doing? Cold Spring Harbor Perspectives in Biology, 11(10), a034728–a034728. https://doi.org/10.1101/cshperspect.a034728.(2018). [↩]
- X. Li, Y. Yang, F. Zhou, Y, Zhang, H. Lu, Q. Jin, L. Gao. SLC11A1 (NRAMP1) Polymorphisms and Tuberculosis Susceptibility: Updated Systematic Review and Meta-Analysis. PLoS ONE, 6(1), e15831–e15831. https://doi.org/10.1371/journal.pone.0015831.(2011). [↩]
- Q. Meilang, Y. Zhang, J. Zhang, Y. Zhao, C. Tian, J. Huang, H. Fan. Polymorphisms in the SLC11A1 gene and tuberculosis risk: a meta-analysis update [Review article]. The International Journal of Tuberculosis and Lung Disease, 16(4), 437–446. https://doi.org/10.5588/ijtld.10.0743.(2012). [↩] [↩]
- Y. Liu, E. Zhao, L. Zhu, D. Zhang, Z. Wang. 3’UTR polymorphisms in NRAMP1 are associated with the susceptibility to pulmonary tuberculosis: A MOOSE-compliant meta-analysis. Medicine, 98(23), e15955–e15955. https://doi.org/10.1097/md.0000000000015955. (2019). [↩] [↩]
- M. K. Stagas, G. S. Papaetis, D. Orphanidou, C. Kostopoulos, S. Syriou, M. Reczko, N. Drakoulis. Polymorphisms of the NRAMP1 gene: Distribution and susceptibility to the development of pulmonary tuberculosis in the Greek population. Medical Science Monitor, 17(1), PH1–PH6. https://doi.org/10.12659/msm.881312.(2011). [↩]
- N. Najmi, G. Kaur, S. K. Sharma, N. K. Mehra. Human Toll-like receptor 4 polymorphismsTLR4Asp299Gly and Thr399Ile influence susceptibility and severity of pulmonary tuberculosis in the Asian Indian population. Tissue Antigens. https://doi.org/10.1111/j.1399-0039.2010.01481.x.(2010). [↩]
- P. J. Martens, C. Gysemans, A. Verstuyf, C. Mathieu.Vitamin D’s Effect on Immune Function. Nutrients, 12(5), 1248–1248. https://doi.org/10.3390/nu12051248. (2020). [↩]
- E. Arrieta-Bolaños, D. Hernández-Zaragoza, R. Barquera. An HLA map of the world: A comparison of HLA frequencies in 200 worldwide populations reveals diverse patterns for class I and class II. Frontiers in Genetics, 14. https://doi.org/10.3389/fgene.2023.866407.(2023). [↩] [↩] [↩]
- J. Dhaliwal, M. Shahnaz, C. Too, A. Azrena, L. Maiselamah, Y. Lee, Y. Irda, M. Salawati. HLA-A, -B and -DR Allele and Haplo- type Frequencies in Malays. ASIAN PACIFIC JOURNAL of ALLERGY and IMMUNOLOGY, 25, 47–51. https://www.apjai-journal.org/wp-content/uploads/2018/01/7HLAABandDRAlleleVol25No1March2007P47.pdf. (2007). [↩]
- Y. Zhang, T. Jiang, X. Yang, Y. Xue, C. Wang, J. Liu, X. Zhang, Z. Chen, M. Zhao, J.C. Li. Toll-Like Receptor -1, -2, and -6 Polymorphisms and Pulmonary Tuberculosis Susceptibility: A Systematic Review and Meta-Analysis. PLoS ONE, 8(5), e63357–e63357. https://doi.org/10.1371/journal.pone.0063357.(2013). [↩] [↩]
- N. Mehra, R.Rajalingam, D. Mitra, V. Taneja, M. Giphart. Variants of HLA-DR2/DR51 Group Haplotypes and Susceptibility to Tuberculoid Leprosy and Pulmonary Tuberculosis in Asian Indians’. 63(2). http://ila.ilsl.br/pdfs/v63n2a06.pdf. (1995). [↩]
- P. J. Martens, C. Gysemans, A. Verstuyf, C. Mathieu.Vitamin D’s Effect on Immune Function. Nutrients, 12(5), 1248–1248. https://doi.org/10.3390/nu12051248. (2020). [↩]
- U. Yadav, P. Kumar, V. Rai. FokI polymorphism of the vitamin D receptor (VDR) gene and susceptibility to tuberculosis: Evidence through a meta-analysis. Infection Genetics and Evolution, 92, 104871–104871. https://doi.org/10.1016/j.meegid.2021.104871. (2021). [↩] [↩]
- Y. Cao, X. Wang, Z. Cao, X. Cheng.Vitamin D receptor gene FokI polymorphisms and tuberculosis susceptibility: a meta-analysis. Archives of Medical Science, 5, 1118–1134. https://doi.org/10.5114/aoms.2016.60092. (2016). [↩]
- Y. Cao, X. Wang, Z. Cao, X. Cheng.Vitamin D receptor gene FokI polymorphisms and tuberculosis susceptibility: a meta-analysis. Archives of Medical Science, 5, 1118–1134. https://doi.org/10.5114/aoms.2016.60092. (2016). [↩]
- T. Kaisho, & S. Akira. Toll-like receptor function and signaling. Journal of Allergy and Clinical Immunology, 117(5), 979–987. https://doi.org/10.1016/j.jaci.2006.02.023.(2024, March 20). [↩]
- P. Selvaraj, M. Harishankar, B. Singh, M.S. Jawahar, V.V. Banurekha. Toll-like receptor and TIRAP gene polymorphisms in pulmonary tuberculosis patients of South India. Tuberculosis, 90(5), 306–310. https://doi.org/10.1016/j.tube.2010.08.001.(2010). [↩]
- M. Mutunga, S.Frison, M. Rava, P. Bahwere. The Forgotten Agenda of Wasting in Southeast Asia: Burden, Determinants and Overlap with Stunting: A Review of Nationally Representative Cross-Sectional Demographic and Health Surveys in Six Countries. Nutrients, 12(2), 559–559. https://doi.org/10.3390/nu12020559 (2020). [↩]
- Q. Meilang, Y. Zhang, J. Zhang, Y. Zhao, C. Tian, J. Huang, H. Fan. Polymorphisms in the SLC11A1 gene and tuberculosis risk: a meta-analysis update [Review article]. The International Journal of Tuberculosis and Lung Disease, 16(4), 437–446. https://doi.org/10.5588/ijtld.10.0743.(2012). [↩]
- N. Mehra, R.Rajalingam, D. Mitra, V. Taneja, M. Giphart. Variants of HLA-DR2/DR51 Group Haplotypes and Susceptibility to Tuberculoid Leprosy and Pulmonary Tuberculosis in Asian Indians’. 63(2). http://ila.ilsl.br/pdfs/v63n2a06.pdf. (1995). [↩]