Abstract
Intracranial hemorrhage (ICH), a life-threatening condition caused by ruptured blood vessels in the brain, is a major contributor to mortality and morbidity worldwide. Elevated intracranial pressure (ICP), a common consequence of ICH, often precedes clinical symptoms, underscoring its importance in detection. Traditional methods for ICP monitoring include catheter-based techniques, which can carry procedural risks. Retinal Imaging has emerged as a non-invasive alternative, offering new biomarkers for early detection, especially Papilledema.Unlike previous AI-based retinal studies that rely on complex deep learning models, this work demonstrates simple, interpretable machine learning algorithms can achieve comparable accuracy for papilledema detection offering a scalable approach for early ICH triage. By utilizing publicly available datasets and correlating retinal biomarkers with clinical outcomes, this research aims to introduce which model could potentially be used as a scalable, cost-effective tool for early ICH diagnosis, reducing mortality and alleviating healthcare burdens globally.
Keywords: Intracranial Hemorrhage (ICH); Papilledema; Retinal Imaging; Fundus Camera; Intracranial Pressure (ICP); Machine Learning (ML); Artificial Intelligence (AI)
Introduction
Fundus photography, an imaging technique to take photos of the back of the eye, has a rich history dating back to the 1800s.1 After the introduction of digital computation in the 1960s and the rise of machine learning, ophthalmic imaging has become a key diagnostic tool across medicine.2 Recent advances in artificial intelligence (AI) now enable non-invasive screening of systemic and neurological diseases directly from retinal images even before noticeable symptoms.3‘4
The retina presents a remarkable ability to serve as an extension to the central nervous system, earning it the term ‘window to the brain’5. As a non-invasive tool, retinal imaging holds high value in monitoring threatening neurological conditions. Without crossing the blood-brain-barrier, an algorithm has the ability to give valuable and timely information saving patients’ lives throughout the world. Current technologies such as Optical Coherence Tomography enhanced with AI, are used to review biomarkers found in neurological diseases. This growth in retinal imaging-based AI models for neurological ailments, is key in detecting Intracerebral Hemorrhage (ICH).
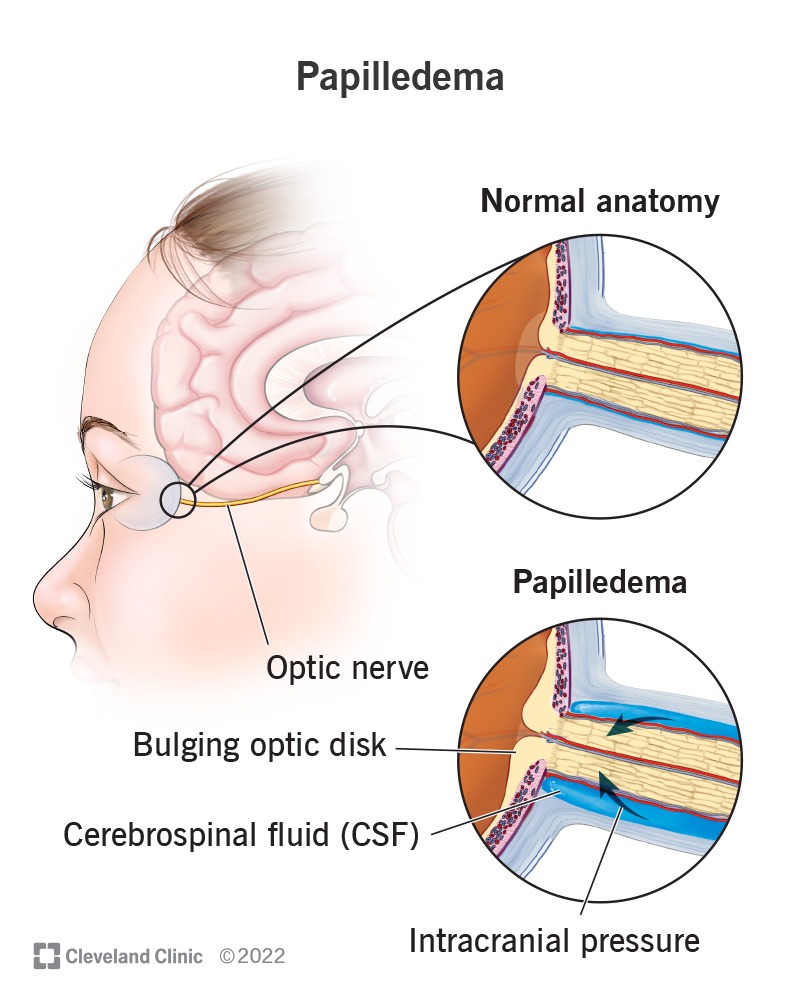
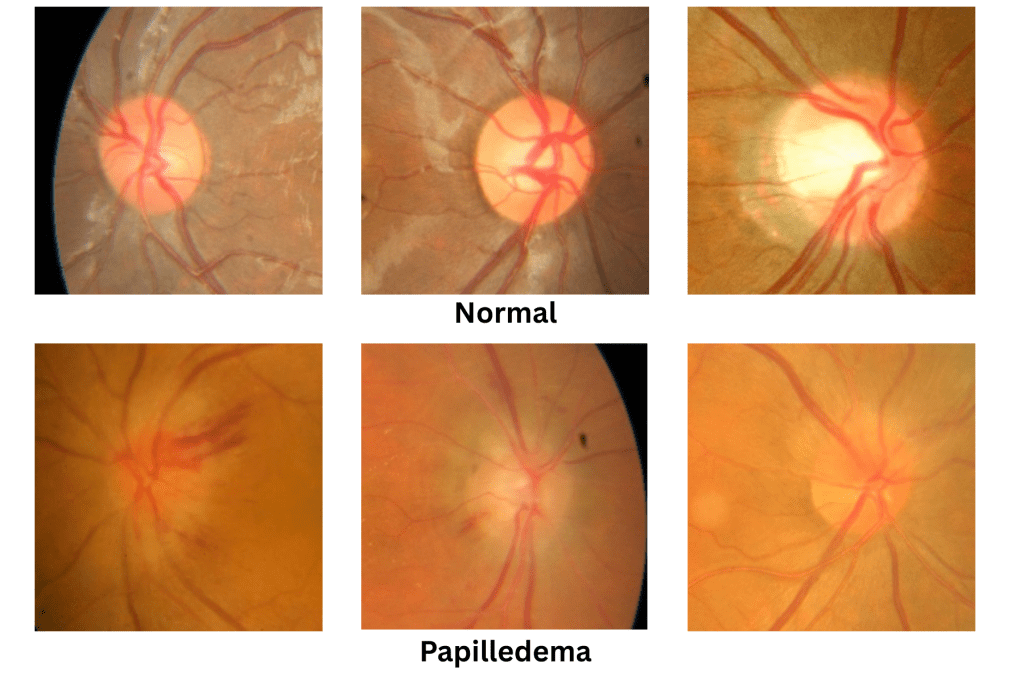
An Intracerebral Hemorrhage occurs when blood bleeds into the brain tissue or between the brain and the skull forming a hematoma, or blood clot, which places pressure onto areas of the brain leading to deadly damage. To date, deaths across the world due to ICH have spiked from 2.3 million in 1990 to 3.3 million in 2021, and this number only continues to grow claiming more lives each year.7 The most dangerous aspect of ICH is the common delay of crucial treatment for a patient, as its symptoms are often not displayed during initial onset. Given the sheer impact of this deadly disease it is crucial to focus our efforts on rapid early detection.8‘9 A key biomarker of Intracerebral Hemorrhage includes a rapid growth in Intracranial Pressure (ICP), increased ICP can place pressure on the optic nerve leading to a physical manifestation in a retinal scan also known as Papilledema [Figure 1].10
Despite advances in AI-assisted ophthalmology, most prior research has focused on diseases like diabetic retinopathy and other ophthalmic disorders, just starting to scratch the surface of predicting ICH risk. Deep-learning models have shown strong performance in papilledema detection, however they require large datasets with extensive preprocessing and significant computational resources, limiting their accessibility in low-resource or emergency settings.11
However, few studies have directly explored using papilledema-based retinal imaging for intracranial hemorrhage triage using lightweight, interpretable machine learning models.12 This study addresses this gap by evaluating the performance of three machine learning algorithms, Logistic Regression, Random Forest, and Support Vector Machines, to classify papilledema from retinal fundus images. By comparing these models’ ability to detect papilledema, the study investigates whether these computationally efficient models can serve as a scalable and cost-effective tool for early detection of elevated ICP and, consequently, intracranial hemorrhage.
Methods
A machine learning model to detect Intracranial Hemorrhages was developed from a dataset of 1,369 colored PNG images, classified into 779 normal images, 295 papilledema images, and 295 pseudo papilledema images [Figure 2]. However for the purpose of this research the pseudo papilledema images were disregarded. Pseudo papilledema is simply an abnormally elevated optic disk without accompanying the optic nerve edema commonly found in Papilledema. From this dataset, the papilledema images found with fundus photography, presents itself as retinal nerve swelling and blurred optic disk. This exclusion was intended to establish a baseline model performance for true pathology detection. A future extension of this work will include both papilledema and pseudo-papilledema to evaluate clinical differentiation in a three-class classification task. Demographic information such as age, sex, and ethnicity was not available in the dataset documentation. The absence of these attributes limits the ability to evaluate model generalizability across diverse populations, and this limitation is explicitly acknowledged in the discussion.
All analyses were performed using Python 3.11. The following open-source Python libraries were used: NumPy, pandas, scikit-learn, and Pillow for image preprocessing model training, and evaluation. This study explores three different models: logistic regression, random forest, and support vector machines (SVM). In every model each image, originally with a minimum size of (208, 240) and maximum size of (240, 240) was converted into grayscale and resized (64×64) to simplify analysis in order to focus on essential features. Two datasets were prepared for comparison: a binary dataset, where pixel intensities were set to 0 or 1, and a grayscale dataset, where continuous intensity values remained. The binary version was used to test whether the structural information alone, such as optic disk or swelling, was enough for classification while the grayscale version retained features that might capture subtle differences. Labelled images were then randomly grouped into an 80-20 ratio for a training and test set for reproducibility. To evaluate generalization stability, 5-fold and 10-fold stratified cross-validation were performed.
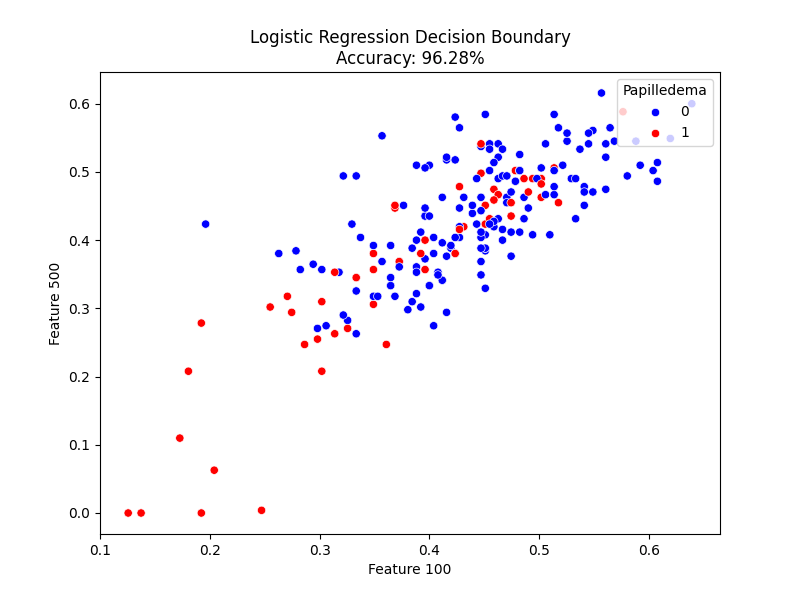
Logistic regression had many appealing qualities over other machine learning algorithms. Its usefulness lies when predicting classification from categories (yes/no, 0/1), and containing a significantly less sensitivity to outliers among other things. In this study, pixel 100 and pixel 500 were selected arbitrarily to generate two-dimensional scatter plots illustrating how the logistic regression model separated the two classes. These pixels were chosen just to help visualize the model and do not represent the same anatomical spot in each image. The model itself used all 4,096 pixels per image during training.
The Random Forest model is a machine learning model that utilizes multiple decision trees, ranging from hundreds to thousands, in order to reach a conclusive result. This model used 100 decision trees each given a random subset of data, in order to add diversity and increase accuracy and reliability in the result. An essential aspect of Random Forest lies in its ability to employ feature importance, for this model we selected the top 10 most important features to display.
Another machine learning algorithm used for classification in this research was, the Support Vector Machines. This model functions by finding a hyperplane to separate the two categories. The model tries to find the optimal hyperplane in order to distance the margin between the two categories. SVM, similar to Logistic Regression, was chosen for its ability to ignore outliers with great effectiveness.
Results
Three models were used to evaluate a machine learning model’s ability to detect intracranial hemorrhages using retinal fundus images. By monitoring the biomarker of papilledema, this study allowed the machine learning model to then detect increased intracranial pressure, a common precursor to the deadly intracranial hemorrhage.
Logistic Regression achieved the highest classification accuracy among all the tested models, with an accuracy of 96.28% [Figure 3]. Even using a simple linear separation between classes, the model clearly distinguished optic disc swelling patterns characteristic of papilledema, demonstrating that simple linear relationships capture meaningful retinal structure differences. The model performed especially well on the normal class with a precision of 0.97 and recall of 0.98. For the papilledema class, its precision reached 0.94 and a recall of 0.91, resulting in an overall macro-averaged F1-score of 0.95 [Figure 7].
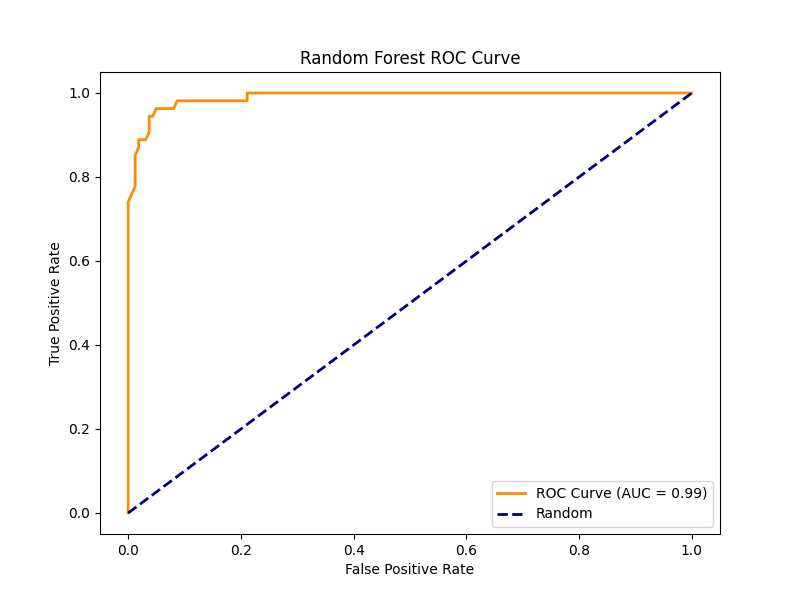
Random Forest achieved an accuracy of 95.81%, similar to SVM. It demonstrated a 0.96 precision and 0.98 recall for normal and 0.94 precision and 0.89 recall for papilledema, showing its comparable classification ability across classes with a macro averaged F1-score of 0.94 [Figure 7].

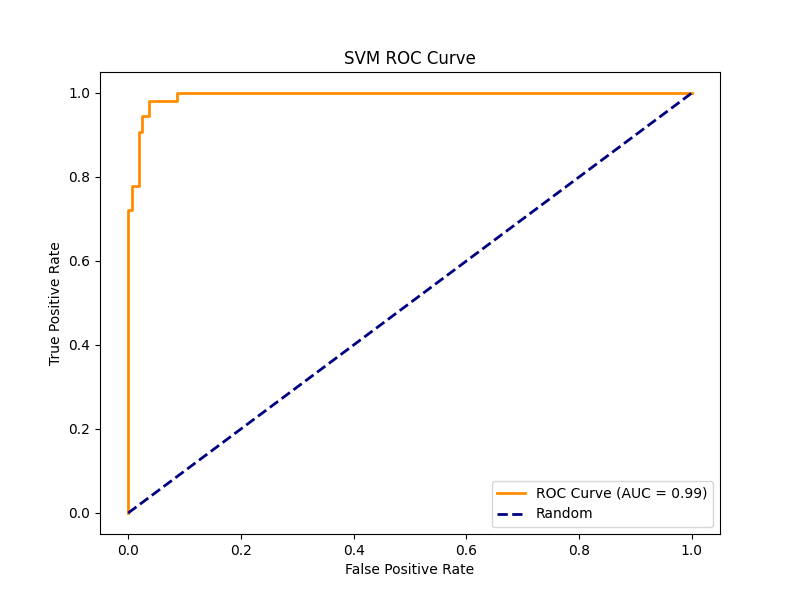
The SVM model gained an overall accuracy of 95.81%. While it had strong performance on the normal class similar to the other models (precision = 0.96, recall = 0.98), it had a lower recall at 0.89 and similar precision at 0.94. The macro-averaged F1-score was 0.94 [Figure 7], proved to be less specific than Logistic Regression.
This study includes a detailed comparison of classification metrics, including precision, recall, F1-score, and support for each class is presented below. This table provides a side-by-side evaluation of each model’s performance on the normal and papilledema classes.

To find the reliability of the reported metrics, using the results from each cross-validation fold 95% confidence intervals were calculated. Logistic Regression achieved a mean 10-fold accuracy of 93.2% (95% CI: 91.6–94.8%), Random Forest reached 93.2% (95% CI: 91.3–95.1%), and SVM got 94.4% (95% CI: 93.3–95.5%). Because the confidence intervals overlap, this proves that the differences in model performance were not statistically significant.
To evaluate image preprocessing, both binary and grayscale datasets were tested. Across all three models, performance differences between grayscale and binary preprocessing were ≤0.5% in accuracy, showing that key structural features were sufficient for reliable classification. This ablation study confirms that the models’ performance was not dependent on fine variations.
Discussion
Unlike multi-center CNN frameworks that require thousands of labeled images, this study demonstrates comparable accuracy using only 1,369 images by simplifying preprocessing and employing interpretable models with reduced computational demand. All three models Logistic Regression, Random Forest, and SVM, are capable of detecting papilledema from retinal images with high accuracy. Among them Logistic Regression had the highest performance, outperforming the other models in accuracy and macro-averaged F1-score. Despite its apparent simplicity, Logistic Regression maintained a highly reliable output, making it a great choice for binary classification in medical imaging.
The simplicity of Logistic Regression makes it an ideal candidate for this application. Simpler models have an added benefit of avoiding overfitting and lack of interpretability, with easier training and faster prediction times, they are more suitable for the analysis of medical images. These efficient models are key in the evolving field of healthcare, and have distinct characteristics that benefit them in various situations.13.
The SVM classifier closely followed, with its excellent ability to model nonlinear boundaries. However its performance with the papilledema class reduced its usefulness, as indicated by a lower recall. Nevertheless, SVM still has key aspects unique to it that allow it to model complex relationships in image data, allowing it to have high specificity and more variety. The Random Forest model matched the SVM in accuracy with the added bonus of feature importance.
The Random Forest feature-importance map [Figure 4] showed that the most predictive pixels were found near the optic disc, the exact region affected first by papilledema when intracranial pressure rises. This confirms that the model used meaningful image regions rather than random pixel noise. The high AUC values in the ROC curves for both Random Forest [Figure 5] and SVM [Figure 6] meaning that the models could correctly identify most papilledema cases while lowering false positives. The Logistic Regression plots and classification summary table also support this, high recall ensures that few true papilledema cases are missed. These insights are key in medical applications, especially with artificial intelligence where explainability is crucial in validating and believing in results.
Despite these promising results, the study acknowledges its limitations, ethical and practical considerations are critical before clinical use. The dataset size, particularly the number of papilledema cases was relatively small compared to the normal dataset, limiting the ability to be able to generalize findings across age, gender, ethnicities, etc. Additionally, variation in image acquisition could introduce noise that affects model consistency. Future research should aim to address these limitations with a larger, more diverse dataset and more selective image collection. While this model cannot directly separate intracranial hemorrhage from other causes of elevated intracranial pressure, it serves as a promising early screening tool to flag potential high-risk patients for further evaluation.
In summary, Logistic Regression emerged as the best model because it offered the highest recall, accuracy, low overfitting, and interpretability. Its performance and transparency make it not only statistically strong but also clinically trustworthy. These characteristics highlight the importance of simple AI in healthcare, where explainability and reproducibility are as important as accuracy itself.
Conclusion
This study has demonstrated the feasibility and effectiveness of using AI algorithms as a tool in diagnosing Intracranial Hemorrhage through early retinal changes. From the models tested, Logistic Regression performed with the highest accuracy overall, underscoring the potential of simple models in clinical diagnosis. While the SVM and Random Forest performed well, offering additional benefits they did not result in a better classification. However, the Random Forest model gave valuable information through feature importance mapping, bringing to light new information of the dataset. By combining non-invasive retinal imaging with machine learning, this approach offers a scalable tool for early Intracranial Hemorrhage detection, especially when neuroimaging methods are unavailable.
To move this work toward real-world application, several next steps are planned. First, the model will be validated on a larger, multi-center dataset that includes patient-level identifiers, allowing for improved generalization. Second, the system will be integrated into a clinical pilot study within hospital emergency departments or neurology clinics to evaluate its usefulness as a screening tool that flags patients for immediate neuroimaging. Third, efforts will focus on developing an automated workflow that combines retinal imaging with basic patient information to provide a rapid “risk score” for elevated intracranial pressure. Finally future work will explore integrating this software as a medical device ensuring it can be used in hospitals and by patients more easily.
Acknowledgements
I want to thank my mentor Ms. Chelsey Beck, for her invaluable support and encouragement throughout this research and to Notre Dame San Jose High School’s Independent Research Program for providing me with this amazing opportunity.
References
- T. J. Bennett. Milestones, rivalries and controversy: The origins of photography and ophthalmic photography – Part III, the first human fundus photograph. History of Ophthalmic Photography Blog, Ophthalmic Photographers’ Society., 2013,https://www.opsweb.org/blogpost/1129727/173791/Milestones-Rivalries-and-Controversy-Part-III. [↩]
- G. Litjens, T. Kooi, B. E. Bejnordi, A. A. A. Setio, F. Ciompi, M. Ghafoorian, J. A. W. M. van der Laak, B. van Ginneken, C. I. Sánchez. A survey on deep learning in medical image analysis. Medical Image Analysis. Vol. 42, pg. 60–88, 2017, https://doi.org/10.1016/j.media.2017.07.005. [↩]
- A. M. Al-Halafi. Applications of artificial intelligence-assisted retinal imaging in systemic diseases: A literature review. Saudi Journal of Ophthalmology. Vol. 37, pg. 185–192, 2023, https://doi.org/10.4103/sjop.sjop_114_22. [↩]
- J. J. Heit, M. Iv, M. Wintermark. Imaging of intracranial hemorrhage. Journal of Stroke. Vol. 19, pg. 11–27, 2017, https://doi.org/10.5853/jos.2016.00563. [↩]
- T. E. Yap, S. I. Balendra, M. T. Almonte, M. F. Cordeiro. Retinal correlates of neurological disorders. Therapeutic Advances in Chronic Disease. Vol. 10, pg. 2040622319882205, 2019, https://doi.org/10.1177/2040622319882205. [↩]
- Cleveland Clinic. Papilledema. Cleveland Clinic Health Library., 2022, https://my.clevelandclinic.org/health/diseases/17817-papilledema. [↩]
- P. Dickens, K. Ramaesh. The evolving role of ophthalmology clinics in screening for early Alzheimer’s disease: A review. Vision (Basel). Vol. 4, pg. 46, 2020, https://doi.org/10.3390/vision4040046. [↩]
- S. Liu, et al. Delayed intracranial hemorrhage after head injury among elderly patients on anticoagulation seen in the emergency department. Canadian Journal of Emergency Medicine. Vol. 24, pg. 853–861, 2022, https://doi.org/10.1007/s43678-022-00351-3. [↩]
- D. Song, D. Xu, M. Li, F. Wang, M. Feng, A. Badr, D. Rigamonti, D. Cistola, D. Yan, J. Zhang, F. Guo. Global, regional, and national burdens of intracerebral hemorrhage and its risk factors from 1990 to 2021. European Journal of Neurology. Vol. 32, pg. e70031, 2025, https://doi.org/10.1111/ene.17003. [↩]
- J. E. Morales-León, L. R. Díaz-De-León. Papilledema in idiopathic intracranial hypertension. Revista Mexicana de Oftalmología. Vol. 97, pg. 99–105, 2023, https://doi.org/10.24875/RMO.22000009. [↩]
- L. Cortés-Ferre, M. A. Gutiérrez-Naranjo, J. J. Egea-Guerrero, S. Pérez-Sánchez, M. Balcerzyk. Deep learning applied to intracranial hemorrhage detection. Journal of Imaging. Vol. 9, pg. 37, 2023, https://doi.org/10.3390/jimaging9020037. [↩]
- D. Milea, et al. Artificial intelligence to detect papilledema from ocular fundus photographs. New England Journal of Medicine. Vol. 382, pg. 1687–1695, 2020, https://doi.org/10.1056/NEJMoa1917130. [↩]
- M. A. Arshad, S. Shahriar, K. Anjum. The power of simplicity: Why simple linear models outperform complex machine learning techniques – case of breast cancer diagnosis. arXiv preprint., 2023, https://doi.org/10.48550/arXiv.2306.02449. [↩]