Abstract
Psychological stress, such as academic overload, increases cortisol levels, which negatively affect skin health. Cortisol modulates the expression of key genes involved in collagen degradation and synthesis, including MMP1, MMP3, and COL1A1. This study investigates how cortisol-induced epigenetic changes, particularly DNA methylation, affect skin cell function. We explored the potential of plant-derived exosomes to reverse these changes and promote skin regeneration. The results suggest that cortisol exposure leads to increased MMP1 and MMP3 expression and decreased COL1A1 expression, impairing collagen synthesis and promoting degradation. Treatment with exosomes derived from Camellia japonica and Rosa spp. (Rosa hybrida) calluses showed a synergistic effect, reducing MMP1 and MMP3 expression while increasing COL1A1 expression. These findings provide insights into the molecular mechanisms of stress-induced skin aging and suggest potential therapeutic applications of exosome-based treatments for skin recovery.
Keywords : Cortisol, Collagen, DNA Methylation, Callus, Epigenetics
Introduction
Psychological stress, which is caused by external pressures such as academic overload, is known to increase systemic cortisol levels, which in turn have a negative impact on skin health. Cortisol is a hormone that plays an important role in the stress response, and while its short-term effects are adaptive, in the long term, high cortisol concentrations have detrimental effects on the skin, such as skin aging, decreased collagen production, and poor tissue regeneration1’2. Recent studies have shown that cortisol modulates the expression of key genes such as MMP1, MMP3, and COL1A1 in skin cells, altering important genes involved in extracellular matrix (ECM) remodeling3’4’5.
Collagen is the most important structural protein in the skin, providing elasticity and support. In particular, COL1A1 is an important gene for collagen synthesis, and its decreased expression is thought to be one of the main causes of skin aging4’6’7. MMP1 and MMP3 are matrix metalloproteinases (MMPs) that play an important role in collagen degradation, and increased expression of these genes promotes collagen degradation and impairs skin regeneration8’9. Thus, the effects of cortisol on the expression of MMP1, MMP3, and COL1A1 have important implications for skin health.
Epigenetic changes, particularly DNA methylation, have been proposed as an important mechanism10 to explain the long-term effects of cortisol. Recent work shows that chronic cortisol exposure induces epigenetic modifications affecting ECM remodeling. In parallel, plant-derived exosome-like nanoparticles have been highlighted for their anti-inflammatory and collagen-protective properties11. Changes in methylation status at gene promoters play an important role in repressing or activating gene expression12’13. Excessive methylation of COL1A1, a collagen synthesis gene, represses its expression and activates the expression of MMP1 and MMP3, collagen degradation genes, which can lead to an imbalance in the skin’s ECM and accelerate skin aging8’4’9. This study aims to explore how cortisol treatment induces DNA methylation in skin cells and how this affects collagen production and degradation.
Conventional approaches to counteract stress-induced skin damage include the use of topical antioxidants and moisturizers, which primarily target oxidative stress and barrier protection14. However, these strategies have limited ability to directly modulate gene expression or epigenetic changes associated with cortisol exposure. In contrast, exosomes offer a unique therapeutic potential by delivering bioactive molecules (such as microRNAs and proteins) that can regulate ECM remodeling and epigenetic pathways at the cellular level.
This study will evaluate the effects of acute and chronic cortisol treatment on the expression and methylation of MMP1, MMP3, and COL1A1 in skin and explore the potential for epigenetic repair using exosomes. By identifying the effects of plant-derived exosomes on the promotion of collagen synthesis and skin regeneration, we aim to provide a potential for the development of novel therapies to reverse stress-induced skin aging.
Methods
Abbreviations
HDF :human dermal fibroblast
EM-seq : Enzymatic-methylation sequencing Rose spp : Rosa hybrida
Cell Culture
Human dermal fibroblasts (HDFs) were obtained from Dr.Kim (Chromogen) and cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin at 37°C in a 5% CO2 incubator.
Cortisol Treatment
Cortisol (Sigma-Aldrich, #C-106) was dissolved in ethanol to prepare stock solutions. Cells were exposed to 1 μM cortisol for acute treatment (1 day) and 1 μM cortisol for chronic treatment (7 days).
Exosome Extraction
Exosomes were derived from Camellia japonica and Rosa spp. calluses. Callus tissues were induced using Murashige and Skoog (MS) medium supplemented with 2,4-dichlorophenoxyacetic acid (2,4-D) and kinetin. After 4 weeks of cultivation, conditioned media were collected and subjected to sequential centrifugation steps (300 × g for 10 min to remove cells, 2,000 × g for 20 min to eliminate debris, and 10,000 × g for 30 min to remove large vesicles). The supernatant was then ultracentrifuged at 100,000 × g for 90 min. Pellets were washed in PBS and re-pelleted at 100,000 × g to reduce protein contamination. To further enrich small extracellular vesicles, a commercial precipitation reagent (ExoQuick, System Biosciences, USA) was applied following the manufacturer’s protocol.
Exosome Characterization
The size and polydispersity index (PDI) of exosomes were determined by dynamic light scattering (DLS) (Malvern Instruments, UK). The presence of exosome markers such as ARF-1 and Hsp70 was confirmed by Western blot analysis using specific antibodies against ARF-1 and Hsp70 (Santa Cruz Biotechnology, USA). The presence of these markers was used to validate the exosome identity and confirm the exosome-enriched fractions.
Gene Expression Analysis
Total RNA was extracted from cells using TRIzol reagent (Invitrogen, USA), and complementary DNA (cDNA) was synthesized using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, USA). Quantitative RT-qPCR was performed to assess the mRNA expression levels of MMP1, MMP3, and COL1A1 using the SYBR Green Master Mix (Takara, Japan) and specific primers for each gene. Quantitative real-time PCR (qPCR) was performed using SYBR Green chemistry (Applied Biosystems). Primer specificity was confirmed by melt-curve analysis, and primer efficiency (90–110%) was validated using standard dilution curves. GAPDH was used as the reference gene. Relative expression levels were calculated using the 2^-ΔΔCt method, and data are presented as mean ± SD from at least three independent biological replicates. MMP1_For : TCCCAAAATCCTGTCCAGCC, MMP1_Rev : CCGGACTTCATCTCTGTCGG, MMP3_For : CCTGGAAATGTTTTGGCCCA, MMP3_Rev : TCATCTTGAGACAGGCGGAA, COL1A1_For : GCTACTACCGGGCTGATGAT, COL1A1_Rev : ACCAGTCTCCATGTTGCAGA
Procollagen Production
Procollagen type I levels were measured using a commercial ELISA kit (Company, Catalog No.). Samples were analyzed in duplicate after 1:10 dilution according to the manufacturer’s instructions. Standard curves (0–1000 pg/mL) were generated for each assay, and the coefficient of variation (CV%) across replicates was <10%.
DNA Methylation Analysis
DNA was extracted from cells using the QIAamp DNA Mini Kit (Qiagen, Germany). Methylation analysis was performed using EM-seq (enzymatic methyl-seq, NEB), a high-sensitivity method for detecting DNA methylation at gene promoters. In this method, TET enzymes were used to selectively oxidize methylated cytosines to hydroxymethylated cytosines. The modified DNA was then subjected to PCR amplification, and next-generation sequencing (NGS) was performed to analyze methylation patterns at the promoters of MMP1, MMP3, and COL1A1.
Library construction
Libraries were constructed according to the manufacturer’s protocol and sequenced on an Illumina platform (150 bp paired-end). The average sequencing coverage across target regions was 30× per base. Promoter regions were defined as −1500 to +500 bp relative to the transcription start site (TSS) based on UCSC genome annotation. Data were processed using the Bismark pipeline for alignment and methylation calling, and methylation levels were calculated as the percentage of methylated cytosines at CpG sites. Average promoter methylation levels were summarized as mean percentages for each condition.
Exosome Treatment
Exosomes isolated from Camellia japonica and Rosa spp. calluses were added to the culture medium of cortisol-treated human dermal fibroblasts. Cells were treated with 10 µg/mL of exosomes for 48 hours.
Statistical Analysis
All experiments were performed with at least three independent biological replicates per condition. For quantitative assays (qPCR, ELISA), each biological replicate was further analyzed with two technical replicates. Data were expressed as mean ± standard deviation (SD). Statistical significance was determined using one-way ANOVA followed by Tukey’s post-hoc test. A p-value < 0.05 was considered statistically significant.
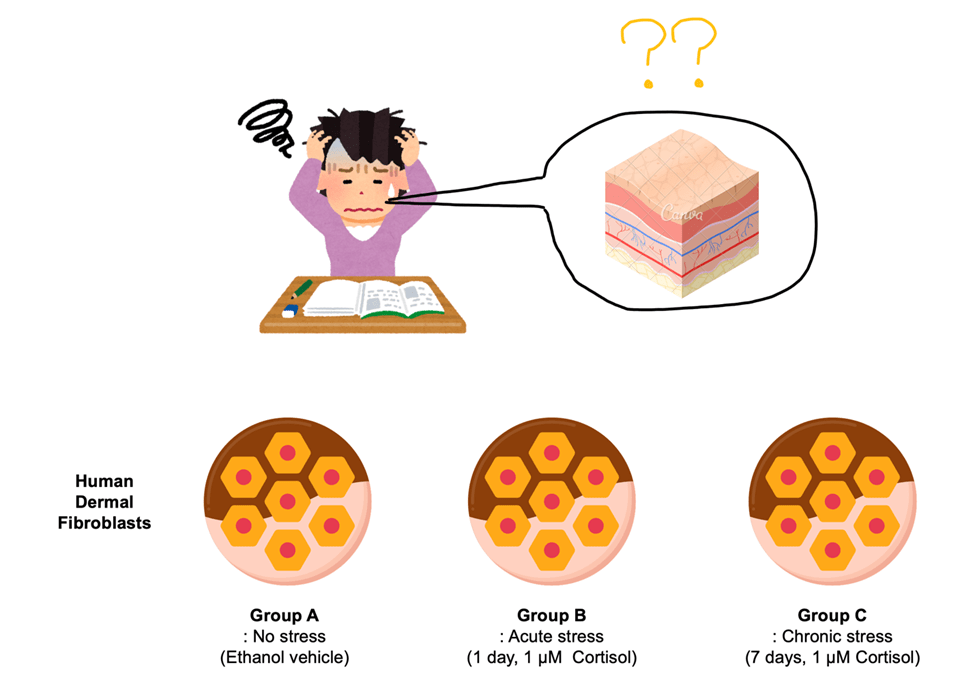
Schematic representation of the experimental hypothesis and workflow. Human skin fibroblasts were subjected to cortisol exposure (acute and chronic)
Psychological stress, such as academic overload, increases systemic cortisol levels, which negatively affect skin health. However, the underlying molecular mechanisms remain unclear. This study hypothesized that cortisol induces DNA hypermethylation in skin cells, leading to altered expression of key structural and remodeling genes, such as COL1A1, MMP1, and MMP3. To test this hypothesis, human dermal fibroblasts were treated with cortisol both acutely (1day, 1 μM) and chronically (7days, 1 μM) (Fig. 1).

(A) RT-qPCR revealed increased MMP1 following cortisol treatment (acute, chronic)
(B) RT-qPCR revealed increased MMP3 following cortisol treatment (acute, chronic)
(C) RT-qPCR revealed decreased COL1A1 following cortisol treatment (acute, chronic)
(D) ELISA confirmed reduced procollagen production following cortisol treatment (acute, chronic)
To evaluate the impact of cortisol treatment on the expression of MMP1, MMP3, COL1A1, and procollagen production in skin cells, RT-qPCR and ELISA experiments were conducted. The results showed a significant increase in the expression of MMP1 and MMP3 after both acute and chronic cortisol treatment, with a sequential increase observed particularly in the chronic treatment group (Fig. 2A and B). This suggests that collagen degradation is promoted. In contrast, the expression of COL1A1 decreased with cortisol treatment, indicating inhibition of collagen synthesis (Fig. 2C). The procollagen ELISA results also showed a significant reduction in procollagen production in both the acute and chronic cortisol-treated groups, reflecting impaired skin regeneration (Fig. 2D). This experiment confirmed that cortisol induces both collagen degradation and inhibition of collagen synthesis in skin cells, negatively affecting skin health. These findings provide important insights into the mechanisms by which stress accelerates skin aging, offering potential avenues for stress management and the development of skin recovery therapies.
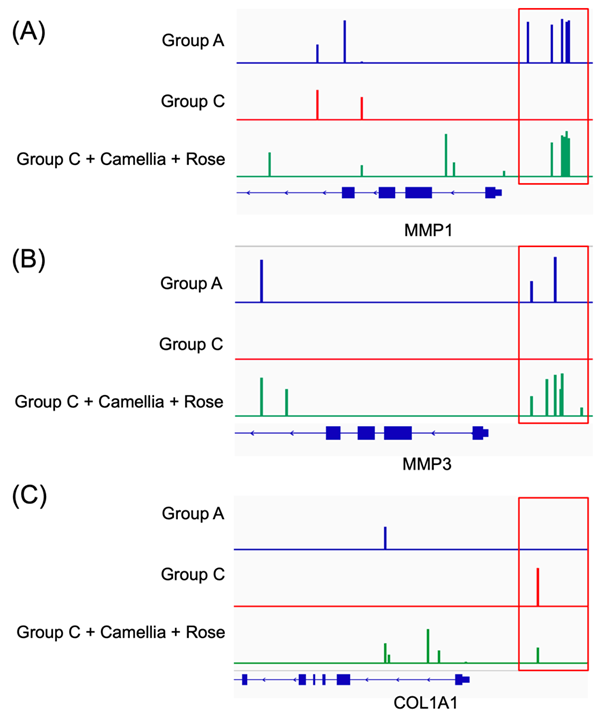
(A) EM-seq analysis showed hypomethylation of MMP1 consistent with gene expression changes induced by chronic cortisol treatment
(B) EM-seq analysis showed hypomethylation of MMP3 consistent with gene expression changes induced by chronic cortisol treatment
(C) EM-seq analysis showed hypermethylation of COL1A1 consistent with gene expression changes induced by chronic cortisol treatment
(D) DNA methylation levels of promoter regions in MMP1, MMP3, and COL1A1.Promoter regions were defined as −1500 to +500 bp relative to the transcription start site (TSS). Average methylation percentages were calculated from EM-seq data.
Epigenetics is an important field that explains how environmental stimuli and stress influence gene function through gene expression regulation mechanisms, such as DNA methylation. Methylation is a key epigenetic mechanism that regulates gene expression, and changes in the methylation status of promoter regions can induce gene silencing or activation. In this study, we used EM-seq (enzymatic methyl-seq) to precisely analyze DNA methylation. EM-seq is a high-sensitivity technique that allows for the detailed assessment of DNA methylation status, making it an extremely useful tool for understanding how stress-induced changes in gene expression occur at the epigenetic level. The results showed that the promoters of MMP1 and MMP3 shifted from hypermethylation to hypomethylation (Figs. 3A and B), which may be associated with increased gene expression. In contrast, the COL1A1 promoter shifted from hypomethylation to hypermethylation (Fig. 3C), which may be related to the inhibition of collagen synthesis. Promoter regions were defined as −1500 to +500 bp relative to the TSS for MMP1, MMP3, and COL1A1. Average methylation percentages within these promoter regions increased under cortisol treatment (MMP1: ~45% vs ~5%; MMP3: ~40% vs ~5%; COL1A1: ~5% vs ~30%), suggesting stress-induced epigenetic alterations in ECM-regulating genes (Fig. 3D). These methylation changes provide important insights into how stress-induced gene expression alterations occur and offer useful data to confirm the potential for epigenetic recovery.

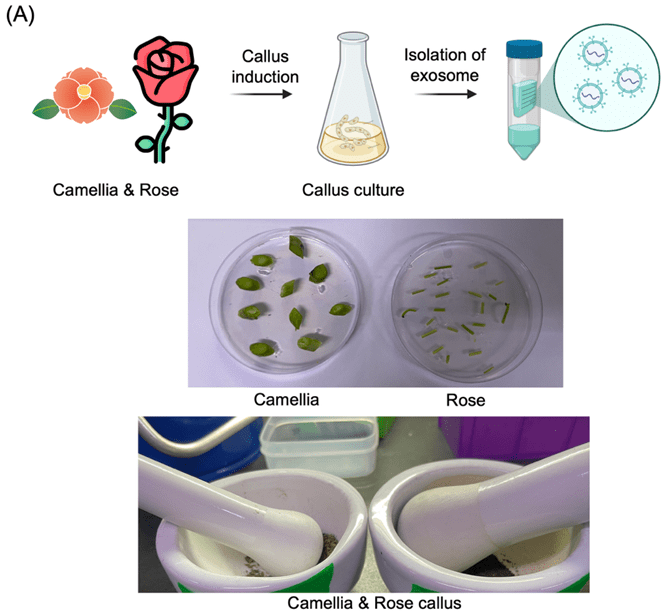
(A) Schematic overview of the exosome extraction process from Camellia and Rosa callus tissues. Callus was induced and cultured for exosome isolation. The lower panel shows mechanical disruption of callus tissues during processing.
(B) Western blot analysis of exosomal markers ARF-1 and Hsp70 in callus tissues (C) and their culture media (M). Both markers were strongly detected in the callus tissue but not in the media.
(C) Experimental treatment groups: Case 1 (Group C only), Case 2 (Group C + Camellia callus-derived exosomes), Case 3 (Group C + Rosa callus-derived exosomes), and Case 4 (Group C + combined Camellia and Rosa callus-derived exosomes).
(D–F) Expression levels of MMP1 (D), MMP3 (E), and COL1A1 (F) assessed by qRT-PCR. Case 4 exhibited the greatest suppression of MMP1 and MMP3 expression and the highest induction of COL1A1 expression. Statistical analysis was performed using one-way ANOVA followed by Tukey’s post-hoc test (*p < 0.05; **p < 0.01; ***p < 0.001).
There have been reports suggesting that plant-derived exosomes can be effective in skin recovery and collagen synthesis promotion15. In this study, callus tissues from Camellia japonica and Rosa spp. were induced, and exosomes extracted from these tissues (Figs. 4A and B) were applied to skin cells to evaluate changes in the expression of MMP1, MMP3, and COL1A1. The presence of exosomes was confirmed by Western blot analysis using specific markers such as ARF-1 and Hsp70 (Fig. 4B), both of which are widely recognized as conserved plant extracellular vesicle markers16, thereby validating the exosome-enriched fractions. The experimental groups included Camellia japonica callus alone, Rosa spp. callus alone, a combined treatment with both exosomes, as well as vehicle-treated controls and vehicle + both exosomes groups (Fig. 4C). The results showed that the expression of MMP1 and MMP3 decreased more significantly in the combined treatment group than in the single treatment groups (Figs. 4D and E), indicating a potential inhibition of collagen degradation. In contrast, the expression of COL1A1 increased synergistically in the combined treatment group (Fig. 4F), suggesting a potential promotion of collagen synthesis. Vehicle-treated and vehicle + exosome groups did not show significant differences compared with untreated controls, supporting the specificity of the observed effects. This experiment demonstrates the synergistic effect of exosomes derived from two plant calluses, and through the decrease in MMP1 and MMP3 expression and the increase in COL1A1 expression, highlights the potential for skin regeneration and skin aging recovery.
(A) EM-seq demonstrated increased methylation at the promoters of MMP1.
(B) EM-seq demonstrated increased methylation at the promoters of MMP3.
(C) EM-seq demonstrated decreased methylation at the promoters of COL1A1.
(D) DNA methylation levels of promoter regions in MMP1, MMP3, and COL1A1.Promoter regions were defined as −1500 to +500 bp relative to the transcription start site (TSS). Average methylation percentages were calculated from EM-seq data.
After applying a combination of Camellia japonica and Rosa spp. callus-derived exosomes, EM-seq was used to analyze the methylation patterns of the promoters of MMP1, MMP3, and COL1A1. The results showed that the promoters of MMP1 and MMP3 shifted to hypermethylation (Figs. 5A and B), resembling the Group A condition. This suggests the recovery of gene silencing. In contrast, the COL1A1 promoter shifted to hypomethylation (Fig. 5C), indicating an epigenetic change that promotes collagen synthesis. This suggests that the combination of exosome treatments acted to activate COL1A1, a key gene in collagen synthesis, thereby potentially contributing to skin regeneration and aging recovery. Furthermore, the combined exosome treatment balanced the expression of MMP1, MMP3, and COL1A1, showing the potential for reversible dual solutions for skin aging regulation.
Discussions
In this study, we utilized EM-seq (Enzymatic Methyl-seq) to analyze the DNA methylation changes induced by cortisol in skin cells. EM-seq is a high-sensitivity technique that uses specific enzymes such as the TET enzyme to selectively modify methylated DNA, thereby enabling efficient and cost-effective assessment of methylation patterns compared to traditional bisulfite sequencing17. By analyzing the promoter methylation status of MMP1, MMP3, and COL1A1, we demonstrated that cortisol regulates genes critical for extracellular matrix (ECM) remodeling and skin health through epigenetic mechanisms.
A key aspect of this study is the use of plant-derived exosomes Callus tissues from Camellia japonica and Rosa spp. were selected given their well-documented skin-beneficial properties, including antioxidant, anti-inflammatory, and collagen-enhancing activities18’19. Exosomes isolated from these calluses reduced the expression of MMP1 and MMP3 while increasing COL1A1expression, with the combined treatment exerting stronger effects than either exosome alone, suggesting a synergistic action. These findings align with recent reports that cortisol alters DNA methylation and ECM homeostasis, while plant exosomes modulate MMP activity and support collagen synthesis20’21.
The combined exosome treatment demonstrated a synergistic effect, where the expression of MMP1 and MMP3 decreased, while the expression of COL1A1 increased. This suggests that the combined exosomes effectively promote collagen synthesis and skin regeneration. The results from Rosa spp. and Camellia japonica callus-derived exosomes further support the positive impact of these exosomes on skin health. Our results are consistent with recent findings that cortisol alters DNA methylation and ECM homeostasis22’23. Moreover, plant exosomes have been reported to modulate MMP activity and support collagen synthesis, supporting their potential as therapeutic agents.
Although the current study demonstrates beneficial effects of plant exosomes on cortisol-induced ECM alterations, the underlying mechanisms require further exploration. Future studies should investigate whether plant exosomes deliver bioactive microRNAs or proteins that directly influence DNA methylation and chromatin regulation in skin fibroblasts. Such mechanisms have been reported in mammalian EVs24, and similar pathways may explain how plant exosomes modulate stress-responsive and ECM-related genes. Functional studies using RNA sequencing of exosome cargo, followed by gain- and loss-of-function assays in recipient cells, would provide mechanistic insights into epigenetic modulation.
Several limitations of this study should be acknowledged. First, we were unable to perform nanoparticle tracking analysis (NTA) or transmission electron microscopy (TEM), which limits the quantitative assessment of particle concentration and morphology. Second, the experiments were performed in vitro, and in vivo validation is required to confirm biological relevance. Follow-up studies using animal skin models would help evaluate exosome penetration, biodistribution, and efficacy under physiological conditions. Third, the scalability of plant exosome production, batch-to-batch consistency, and quality control remain challenges. Optimizing bioreactor-based callus culture systems and developing standardized purification pipelines will be critical for translational application. Third, the immunogenicity and delivery methods of plant exosomes to human skin require careful evaluation to ensure safety. Lastly, while preliminary product prototyping was attempted, rigorous clinical-grade testing is essential before any therapeutic or cosmetic application.
In conclusion, this study demonstrates that Camellia japonica and Rosa spp. callus-derived exosomes modulate cortisol-induced ECM changes through DNA methylation and synergistic regulation of collagen metabolism. Future mechanistic studies on microRNA/protein cargo and comprehensive in vivo validation will be crucial for advancing plant exosomes as promising candidates for anti-aging skin therapies.
Ethics/Material Statement
This study did not involve human participants or animal subjects. All experiments were performed using plant callus tissues (Camellia japonica and Rosa hybrida) and human cell lines (HDFs) obtained from Korea Cell Line Bank, following relevant institutional biosafety guidelines.
References
- Chen Y, Lyga J. Brain-skin connection: stress, inflammation and skin aging. Inflamm Allergy Drug Targets. 13(3), 177-90 (2014), Chae M, Bae IH, Lim SH, Jung K, Roh J, Kim W. AP Collagen Peptides Prevent Cortisol-Induced Decrease of Collagen Type I in Human Dermal Fibroblasts. Int J Mol Sci. 22(9):4788 (2021) [↩]
- Vukelic, S., Stojadinovic, O., Pastar, I., Rabach, M., Krzyzanowska, A., Lebrun, E., Davis, S. C., Resnik, S., Brem, H., Tomic-Canic, M. Cortisol synthesis in epidermis is induced by IL-1 and tissue injury. J Biol Chem 286, 10265-10275 (2011). [↩]
- Chen Y, Lyga J. Brain-skin connection: stress, inflammation and skin aging. Inflamm Allergy Drug Targets. 13(3), 177-90 (2014) [↩]
- Yang EV, Bane CM, MacCallum RC, Kiecolt-Glaser JK, Malarkey WB, Glaser R. Stress-related modulation of matrix metalloproteinase expression. J Neuroimmunol. 133(1-2) (2002) [↩] [↩] [↩]
- Smith, J.C., Boone, B.E., Opalenik, S.R., Williams, S.M., Russell, S.B. Gene profiling of keloid fibroblasts shows altered expression in multiple fibrosis-associated pathways. J Invest Dermatol 128, 1298-1310 (2008). [↩]
- Bigot, N., Beauchef, G., Hervieu, M., Oddos, T., Demoor, M., Boumediene, K., Galéra, P. NF-κB Accumulation Associated with COL1A1 Transactivators Defects during Chronological Aging Represses Type I Collagen Expression through a –112/–61-bp Region of the COL1A1 Promoter in Human Skin Fibroblasts. Journal of Investigative Dermatology 132, 2360-2367 (2012) [↩]
- Lago, J.C., Puzzi, M.B. The effect of aging in primary human dermal fibroblasts. PLoS One 14, e0219165 (2019). [↩]
- Chae M, Bae IH, Lim SH, Jung K, Roh J, Kim W. AP Collagen Peptides Prevent Cortisol-Induced Decrease of Collagen Type I in Human Dermal Fibroblasts. Int J Mol Sci. 22(9):4788 (2021) [↩] [↩]
- Xia, W., Hammerberg, C., Li, Y., He, T., Quan, T., Voorhees, J. J., & Fisher, G. J. Expression of catalytically active matrix metalloproteinase-1 in dermal fibroblasts induces collagen fragmentation and functional alterations that resemble aged human skin. Aging Cell 12, 661-671 (2013). [↩] [↩]
- Gopinathan, G., Diekwisch, T.G.H. Epigenetics and Early Development. J Dev Biol 10(2022). [↩]
- Zhao, B., Lin, H., Jiang, X., Li, W., Gao, Y., Li, M., Yu, Y., Chen, N., & Gao, J. Exosome-like nanoparticles derived from fruits, vegetables, and herbs: innovative strategies of therapeutic and drug delivery. Theranostics 14, 4598-4621 (2024). [↩]
- Wilkinson, M.F. Evidence that DNA methylation engenders dynamic gene regulation. Proc Natl Acad Sci U S A 112, E2116 (2015) [↩]
- Dhar, G.A., Saha, S., Mitra, P., Nag Chaudhuri, R. DNA methylation and regulation of gene expression: Guardian of our health. Nucleus (Calcutta) 64, 259-270 (2021). [↩]
- Kim, J.H., Kwack, M.H., Lee, W.J. Effects of antioxidants on skin hydration, inflammatory cytokines, and keratinocyte differentiation markers in a PM(10)-exposed skin barrier-disrupted mouse model. Int J Immunopathol Pharmacol 38, 3946320241303860 (2024). [↩]
- Wei W, Ren X, Yi F, Zhang X, Hou J, Zhang Z, Yuan L, Li L, Gao Q. Innovative Plant Exosome Delivery System for Enhancing Antiaging Potency on Skin. ACS Appl Bio Mater. 8(3):2117-2127 (2025). [↩]
- Qian, Z., Shen, Q., Yang, X., Qiu, Y., Zhang, W. The Role of Extracellular Vesicles: An Epigenetic View of the Cancer Microenvironment. Biomed Res Int 2015, 649161 (2015). [↩]
- Olova, N.N., Andrews, S. Whole Genome Methylation Sequencing via Enzymatic Conversion (EM-seq): Protocol, Data Processing, and Analysis. in High Throughput Gene Screening: Methods and Protocols (eds. Carabetta, V.J. & Akintunde, O.) 73-98 (Springer US, New York, NY, 2025), Longtin A, Watowich MM, Sadoughi B, Petersen RM, Brosnan SF, Buetow K, Cai Q; Cayo Biobank Research Unit; Gurven MD, Higham JP, Highland HM, Huang YT, Kaplan H, Kraft TS, Lim YAL, Long J, Melin AD, Montague MJ, Roberson J, Ng KS, Platt ML, Schneider-Crease IA, Stieglitz J, Trumble BC, Venkataraman VV, Wallace IJ, Wu J, Snyder-Mackler N, Jones A, Bick AG, Lea AJ. Cost-effective solutions for high-throughput enzymatic DNA methylation sequencing. PLoS Genet. 21(5):e1011667 (2025). [↩]
- Karabay, A.Z., Barar, J., Hekmatshoar, Y., Rahbar Saadat, Y. Multifaceted Therapeutic Potential of Plant-Derived Exosomes: Immunomodulation, Anticancer, Anti-Aging, Anti-Melanogenesis, Detoxification, and Drug Delivery. Biomolecules 15(2025) [↩]
- Muraca, M., Cappariello, A. The Role of Extracellular Vesicles (EVs) in the Epigenetic Regulation of Bone Metabolism and Osteoporosis. Int J Mol Sci 21(2020). [↩]
- Hsu P, Kamijyo Y, Koike E, Ichikawa S, Zheng Y, Ohno T, Katayama S. Exosome-like nanovesicles derived from kale juice enhance collagen production by downregulating Smad7 in human skin fibroblasts. Front Nutr. 12:1486572 (2025) [↩]
- Bin Dayel S, Hussein RS. Exosomes in Dermatology: Emerging Roles in Skin Health and Disease. Pharmaceutics. 17(5):600 (2025). [↩]
- Nicolaides, N.C., Chrousos, G.P. Glucocorticoid Signaling Pathway: From Bench to Bedside. Int J Mol Sci 24(2023) [↩]
- Leung, C. S., Kosyk, O., Welter, E. M., Dietrich, N., Archer, T. K., Zannas, A. S. Chronic stress-driven glucocorticoid receptor activation programs key cell phenotypes and functional epigenomic patterns in human fibroblasts. iScience 25, 104960 (2022). [↩]
- Urzì, O., Gasparro, R., Ganji, N.R., Alessandro, R., Raimondo, S. Plant-RNA in Extracellular Vesicles: The Secret of Cross-Kingdom Communication. Membranes (Basel) 12(2022). [↩]