Abstract
Background: The Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-Cas9 gene-editing technique has emerged as a significant breakthrough in agricultural biotechnology. As global hunger continues, it is essential to examine how CRISPR-Cas9 applications can assist in ensuring sufficient food production and resource management sustainability.
Objectives: This review examines the application of CRISPR-Cas9 in major staple crops (e.g., rice, wheat, maize, and soybeans), focusing on genetic modifications that improve productivity, nutrition, and environmental stress tolerance. Particularly, this review organizes recent research findings, which is relevant to food-insecure and resource-limited regions.
Methods: We identified and verified peer-reviewed articles from 2014 to 2024 through targeted searches in PubMed, Scopus, and Web of Science. Studies were included if they reported experimental CRISPR-Cas9 interventions in staple crops, provided validated outcomes, and addressed factors potentially related to food security. We extracted and grouped key data points to capture recurring themes in the research focus, then synthesized these findings in a narrative format to highlight overarching themes and trends.
Results: Analyses of the included studies revealed that CRISPR-Cas9 research in major crops predominantly centered around three core themes: (1) disease resistance by editing susceptibility genes, (2) enhanced nutritional content through metabolic pathway alterations, and (3) climate resilience by engineering stress-responsive genes. Studies that focused on CRISPR-Cas9 edited crops showed improvement of β-carotene with its sixfold increase in rice and banana, GABA content increase up to 15-fold in tomato, and drought-tolerance maize yield also increased 5% more under stresses. These results show that CRISPR’s ability to enhance multiple traits is beneficial for crops. Despite potential benefits, there are implementation barriers. Most CRISPR-edited crops are still only tested in laboratory conditions, not real farms. In addition, access to biotechnology and infrastructure remains limited to small-scale farmers in low-resource regions, especially in parts of Africa and Asia, the easier access and to improve their farm condition and increase harvest should be urged.
Keywords: CRISPR-Cas9, gene editing, agricultural biotechnology, food security, global hunger, crop improvement, disease resistance, nutritional enhancement, climate resilience, staple crops, genome modification, sustainable agriculture, developing countries, biotechnology policy, food production.
Introduction
Plant diseases contribute significantly to global food insecurity, which currently affects over 800 million people1. Rice blast disease from the southern US states such as Louisiana, Arkansas, and Mississippi, threatened the nutrition stability of 60 million people. Without the use of fungicides, the loss could rise to $1.1 Billion USD, worsening the economic loss. Also, the drought in sub-Saharan Africa failed to provide agricultural stability2,3.
Though preceding reviews stated that CRISPR-Cas9 has emerged as a transformative tool for crop improvement, they often lacked practical implementation such as field validation, regulatory barriers, or regional infrastructure. This review argues that CRISPR-Cas9 has a potential to challenge biotechnology breakthroughs for nutritional enhancement by introducing genetically modified staple crops to fight disease and climate crisis, with a focus on its practical applications. This review assesses the application of CRISPR-Cas9 to the various regions, particularly focusing on Asia and Africa, where food systems and governance framework for agricultural biotechnology differ.
Global Hunger and Food Security Context
Global hunger is a complex issue closely linked to various aspects of food security. Plant diseases exacerbate food insecurity, affecting approximately 800 million undernourished people worldwide1. Various pathogens—such as fungi, bacteria, viruses, and nematodes—are estimated to reduce global crop yields by 20-40% annually4. For instance, rice blast disease causes annual yield losses that could feed around 60 million people2.
In addition to the challenges posed by pathogens, staple crops often suffer from nutritional deficiencies. While these crops provide essential calories needed for survival, they frequently lack the micronutrients necessary for optimal human health5. Vitamin A deficiency affects millions of children across sub-Saharan Africa, increasing their vulnerability to blindness and severe infections due to compromised immune function6.
Climate change threatens global food security, including rising temperatures, unpredictable rainfall patterns, and increasing extreme weather events. These climate factors not only directly affect crop yields but also contribute to the spread of agricultural pests and diseases7. Recurring drought episodes in sub-Saharan Africa have led to consecutive maize crop failures by diminishing immediate food availability and long-term agricultural stability3.
The Potential of CRISPR-Cas9 Technology
In this review, the term ‘genome editing’ and ‘gene editing’ are used interchangeably when referring to targeted modification of DNA sequences using CRISPR-Cas9. Though the terms are technically different where “genome editing” refers to changes at broader genomic level while “gene editing” refers to specifically targeted genes, the two terms are frequently used and treated equivalently in many studies. Therefore, for the purposes of consistency and clarification, the review adopts “genome editing” as a standard term unless a distinction is necessary.
CRISPR-Cas9 technology represents a groundbreaking advancement in agricultural biotechnology, providing efficiency in genetic modification8. Unlike traditional genetic modification methods, this gene-editing system enables targeted DNA alterations without necessarily integrating foreign genetic material, addressing key concerns often associated with conventional Genetically Modified Organisms (GMO) technologies9.
CRISPR-Cas9 facilitates precise modifications within crop genomes, enhancing desirable traits such as disease resistance, yield improvement, and environmental stress tolerance10. This technology has shown considerable potential to accelerate breeding timelines by introducing targeted genetic changes directly into plant germlines and significantly reducing development time compared to traditional breeding methods11.
CRISPR-Cas9 offers promising solutions for global food security by developing crops that resist pests, diseases, and environmental stresses while improving agricultural productivity and minimizing yield losses12. CRISPR-Cas9 has the potential to transform agricultural practices and support sustainable global food systems13. However, despite its potential, CRISPR-Cas9 applications face notable limitations, including efficiency in some crops, success in field trials, and regulatory uncertainties.
CRISPR-Cas9 and the Sustainable Development Goals (SDGs)
Using CRISPR-Cas9 technology in agriculture is crucial for achieving the United Nations Sustainable Development Goals, particularly SDG 2: Zero Hunger. This underscores its potential to enhance global food security and may contribute to reducing hunger by 203014. The CRISPR-Cas9 supports sustainable agricultural practices through multiple pathways. Crops with improved disease resistance can reduce reliance on chemical pesticides and promote environmentally friendly farming methods. Additionally, improved genetic traits increase crop yields, making land use more efficient and reducing stress on expanding agricultural limits. By increasing tolerance to environmental stressors like drought and nutrient deficiencies, CRISPR-Cas9 fosters more efficient use of natural resources, such as water and soil nutrients11. CRISPR-Cas9 offers promising tools for crop improvement; however, infrastructure challenges and technical limitations remain, especially in translating the technology to the field.
This review aims to compile recent research findings to assess the role of CRISPR-Cas9 in addressing agricultural challenges. This research is to evaluate the effectiveness of gene-editing applications and explore the feasibility of integrating CRISPR-Cas9 into sustainable agricultural practices. Furthermore, given the urgent need to enhance food production in resource-limited regions, this study will also assess how CRISPR-Cas9 is being implemented in developing countries and the obstacles to its adoption. Ultimately, this review intends to provide insights into how gene-editing technologies can contribute to global food security and inform future research directions.
Research Questions
The following research questions will help to direct this review:
- What are the common agricultural traits altered by CRISPR-Cas9 in staple crops from 2014 to 2024?
- What genome-editing technologies are used in improving the resilience and nutritional value of crops?
To address these questions, this review follows a structured approach based on systematic review principles. Studies were selected using defined search criteria, with thematic synthesis across three core domains: disease resistance, nutritional biofortification, and climate resilience.
Methods
This investigation employed a systematic literature search using three major academic databases—PubMed, Scopus, and Web of Science—to identify peer-reviewed experimental studies on CRISPR-Cas9 applications in key food crops, including rice, wheat, maize, and soybean. To highlight geographic differences the regional information on field trials, crop target, and regional regulatory approaches were mentioned. The search was limited to articles published between January 2014 and January 2024 and written in English. The search strategy included combinations of the following terms: “CRISPR-Cas9” OR “gene editing” OR “genome editing” AND “agriculture” OR “crop improvement” OR “food security” OR “staple crops” AND “disease resistance” OR “nutritional enhancement” OR “biofortification” OR “stress tolerance” OR “climate resilience.”. Themes used in this review were considered by single-reviewer analysis through frequency and significance of topics in the literature, which included tolerance on disease, drought, or cold. This method made a comprehensive review, but may lead to biased research that should be reviewed later.
Inclusion Criteria
Studies qualified for inclusion if they provided experimental evidence demonstrating CRISPR-Cas9 genome editing in staple food crops directly relevant to food security challenges. Each included study needed to present validated phenotypic data, such as disease resistance assessments, nutrient composition analyses, or stress tolerance evaluations, rather than focusing exclusively on molecular or theoretical aspects. Only peer-reviewed articles published in recognized academic journals were included. Studies focusing on non-food model plants or published outside the 2014–2024 time frame were excluded from the following evaluation analysis. However, a few studies on non-staple crops–including tomato and citrus–were included when their methods were innovative in CRISPR-Cas9 technologies such as transgene-free editing and promoter modification, which could be also implemented in future research in staple crops. This is why the Liu et al. study was included in this review even though the study did not use CRISPR-Cas9; the mention of the literature has the potential of providing a framework for future reference for the staple crops.
Exclusion Criteria
Studies lacking empirical evidence to support phenotypic changes resulting from CRISPR-Cas9 modifications were excluded. This criterion applies to review papers, theoretical analyses, and opinion articles that do not present original experimental findings. Additionally, research focusing solely on model plants (e.g., Arabidopsis thaliana) without direct relevance to food crops was omitted. Non-English publications and studies lacking full-text availability from reputable scientific databases were also excluded. Duplicate data and non-novel findings were removed to maintain analytical integrity.
Data Extraction and Thematic Synthesis
The full texts of selected studies underwent a thorough review to extract key information using a standardized data collection framework. The extracted details included crop species, target traits, edited genes, and CRISPR-Cas9 methodologies (such as gene knockout, base editing, and promoter modification), along with experimental validation approaches (including pathogen resistance assays, nutrient quantification, and abiotic stress tests) and principal findings related to observed phenotypic effects. We identified common patterns in gene targets, editing strategies, and phenotypic improvements across all categories. Additionally, we analyzed several key challenges in the field, such as off-target mutations, the efficiency of gene transformation, and regulatory issues.
Quality and Reliability Assessment
Studies were assessed for reliability based on the quality of experimental design, including replication, control group methods, and statistical validation. While no studies were excluded solely due to quality concerns, limitations in experimental design, sample size, or validation techniques were noted to inform the discussion of challenges and future directions in CRISPR-Cas9 research for food crops.
Results
CRISPR-Cas9 Applications in Crop Improvement (2014-2024)
Table 1: This table summarizes the applications of CRISPR-Cas9 in crop improvement from 2014 to 2024. It details each study’s reference, crop species, target genes, trait focus, experimental setting, and key outcomes, emphasizing advancements in disease resistance, yield, nutritional quality, and stress tolerance.
| Trait Focus | Crop Species | Target Gene(s) | Setting | Key Outcomes | Study (Reference) |
| Disease resistance | Wheat (Triticum aestivum) | TaMLO (3 homoeologs) against powdery mildew | Greenhouse | Triple MLO knockout in wheat yields heritable powdery mildew resistance with no growth penalties | Wang et al.15 |
| Rice (Oryza sativa) | OsERF922 (knockout) against rice blast | Greenhouse/Field | OsERF922 knockout in rice boosts blast resistance with stable T2 inheritance and no agronomic trade-offs. | Want et al.16 | |
| Tomato (Solanum lycopersicum) | SlMLO1 (48bp deletion) against powdery mildew | Greenhouse | 48bp deletion in SlMLO1 yields transgene-free, stable powdery mildew resistance in tomato with no growth or fruit penalties. | Nekrasov et al.17 | |
| Sweet orange (Citrus sinensis Osbeck) | WRKY22 (transcription factor) against citrus canker | Laboratory/Greenhouse | WRKY22: Negative regulator; upregulates CsLOB1 and cell wall genes, increasing citrus canker susceptibility | Long et al.18 | |
| Rice (Oryza sativa) | OsSWEET genes (promoter edits) against bacterial blight | Field trial | SWEET promoter edits block TAL effectors, yielding durable, broad-spectrum bacterial blight resistance in rice with no yield loss. | Oliva et al.19 | |
| Banana (Musa spp.) | Integrated endogenous banana streak virus (eBSV) sequences | Laboratory (tissue culture) | Inactivated B-genome viral sequences via CRISPR, yielding 75% virus-free bananas under stress with no developmental impact. | Tripathi et al.20 | |
| Cassava (Manihot esculenta) | nCBP-1 and nCBP-2 (eIF4E isoforms) against brown streak | Greenhouse/Field | eIF4E knockout in cassava via CRISPR reduces brown streak severity and virus load, validated in Ugandan fields. | Gomez et al.21 | |
| Citrus (Sweet orange, Citrus sinensis) | CsLOB1 (LATERAL ORGAN BOUNDARIES 1) against citrus canker | Greenhouse | CsLOB1 promoter editing via CRISPR blocks X. citri activation, conferring robust citrus canker resistance. | Jia et al.22 | |
| Wheat (Triticum aestivum) | TaEDR1 (three homoeologs) against powdery mildew | Greenhouse/Laboratory | Triple TaEDR1 knockout in wheat via CRISPR-Cas9 boosts powdery mildew resistance without yield penalties, enabling multiplex editing for fungicide-free breeding. | Zhang et al.23 | |
| Nutritional enhancement | Tomato (Solanum lycopersicum) | SlGAD2 and SlGAD3 (glutamate decarboxylase genes) to increase GABA | Greenhouse | SlGAD2/3 truncation via CRISPR yields up to 15x GABA boost in tomato with no quality trade-offs. | Nonaka et al.24 |
| Soybean (Glycine max) | FAD2-1A & FAD2-1B (double knockout) for oil quality | Greenhouse | Double FAD knockout yields soybean oil with 80% oleic acid, enhancing stability and nutrition. | Do et al.25 | |
| Wheat (Triticum aestivum) | α-gliadin genes to reduce gluten | Greenhouse | α-gliadin mutations lower immunoreactivity by up to 85% while preserving baking quality. | Sánchez-León et al.26 | |
| Wheat (Triticum aestivum) | TaVIT2 (Vacuolar Iron Transporter) for iron biofortification | Laboratory/Greenhouse | TaVIT2 overexpression in wheat endosperm doubles iron in white flour without yield loss, showcasing strong biofortification potential. | Connorton et al.27 | |
| Banana (Musa acuminata) | LCYε (lycopene epsilon-cyclase) to increase β-carotene | Laboratory/Greenhouse | CRISPR-Cas9 knockout of LCYε in banana boosts β-carotene 6-fold, stably inherited with no trait trade-offs. | Kaur et al.28 | |
| Climate resilience | Rice (Oryza sativa) | OsRR22 (knockout) for salt tolerance | Greenhouse/Field | OsRR22 knockout enhances salt tolerance and yield without agronomic penalties. | Zhang et al.29 |
| Rice (Oryza sativa) | OsNAC14 (transcription factor) for drought tolerance | Laboratory/Greenhouse/Field trials | OsNAC14 overexpression boosts rice drought tolerance via stress gene regulation and enhanced root growth. | Shim et al.30 | |
| Soybean (Glycine max) | GmSALT3 for salt tolerance | Laboratory/Field trials | GmSALT3 enhances soybean salt tolerance by excluding Cl⁻, improving photosynthesis and yield under salinity in later growth stages. | Liu et al.31 | |
| Rice (Oryza sativa) | OsGATA16 (transcription factor) and OsWRKY45-1 for cold tolerance | Laboratory/Greenhouse | OsGATA16 boosts rice cold tolerance by repressing OsWRKY45-1, enhancing survival and reducing ROS. | Zhang et al.32 | |
| Maize (Zea mays) | ARGOS8 (promoter replacement) for drought tolerance | Field trial | ARGOS8 promoter swap yields ~5% drought yield boost, stable edits, and reduced ethylene sensitivity. | Shi et al.33 | |
| Yield and quality improvement | Wild tomato (Solanum pimpinellifolium) | Multiple domestication genes | Greenhouse | Six-gene edit in wild tomato: 3x fruit size, 10x fruit number, 500% lycopene increase, and improved architecture. | Zsögön et al.34 |
While the primary focus of this review is staple crops such as rice, wheat, maize, and more, a limited number of studies, including tomato and citrus, were mentioned in the table when they demonstrated methodological advancements in CRISPR-Cas9 application. For instance, transgene-free genome editing was used in tomatoes, and novel promoter-editing strategy was used in citrus35,36.
Enhancing Disease Resistance
This systematic analysis highlighted disease resistance as a primary focus of CRISPR-Cas9 applications in crop improvement research, accounting for a substantial proportion of the studies included in this review. These investigations showcased a variety of gene-editing strategies to combat fungal, bacterial, and viral pathogens across significant staple crops, with several approaches advancing to promising field-level validation.
Fungal Disease Resistance
CRISPR-Cas9 has successfully engineered resistance against powdery mildew, a common fungal disease that affects many crops. In a groundbreaking study, Wang et al. simultaneously targeted all three homeologs of the TaMLO gene in hexaploid bread wheat37. This knockout approach resulted in heritable resistance to powdery mildew without any noticeable growth penalties, marking the first demonstration of CRISPR-Cas9 genome editing in this complex polyploid crop. Nekrasov et al. further validated the effectiveness of targeting MLO genes by creating a 48bp deletion in the SlMLO1 gene of tomato, leading to transgene-free plants with strong resistance to powdery mildew (Oidium neolycopersici)36. This resistance remained stable through T1 and T2 generations, without impacting plant growth or fruit production, underscoring CRISPR-Cas9’s potential for developing non-GMO disease-resistant varieties.
The advancement of targeting multiple homeologous gene copies in polyploid crops continued with Zhang et al.’s simultaneous modification of all three homeologs of TaEDR1 in wheat38. Their research showed that resistance to powdery mildew increased with the number of mutated homeologs, with triple mutants demonstrating the highest resistance.
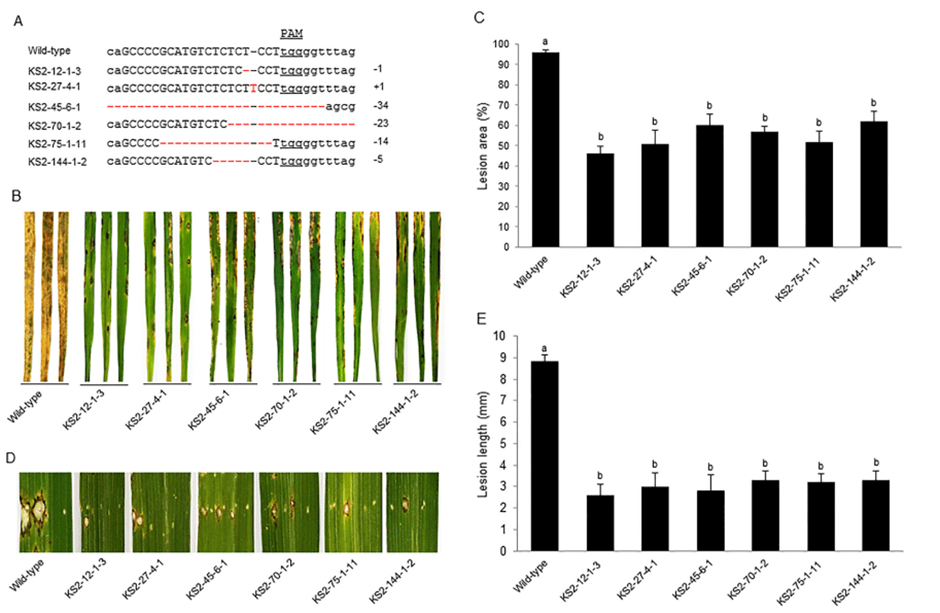
Rice blast disease, caused by Magnaporthe oryzae, poses another significant fungal threat to global food security. Wang et al. tackled this issue by targeting OsERF922, an ethylene response factor in rice39. The CRISPR-mediated knockout of this gene significantly enhanced rice blast resistance with stable inheritance through the T2 generation while preserving typical agronomic characteristics. As illustrated in Panel B and C of Fig. 2 from the original study by Wang et al., CRISPR-edited rice lines exhibited around 40-60% lower lesion area due to fungal disease15. The histograms from Panel C and E effectively highlight that OsERF922 knockout enhanced the fungal disease resistance. Also, Panel B and D depicts the visual symptom reductions in CRISPR-edited lines compared to wildtypes15.
Bacterial Disease Resistance
Bacterial diseases present significant threats to crop production, and CRISPR-Cas9 has enabled innovative approaches to combat them. Researchers have validated two strategies for addressing citrus canker, a devastating bacterial disease affecting citrus crops. Jia et al. targeted the promoter region of the susceptibility gene CsLOB1 in sweet orange, creating mutations in the effector binding element (EBE) recognized by the bacterial pathogen’s transcription activator-like (TAL) effector35. This modification prevented Xanthomonas citri from activating the susceptibility gene, resulting in strong resistance to citrus canker.
Long et al. provided complementary insights by identifying WRKY22 as a negative regulator of citrus canker resistance40. Their research revealed that WRKY22 increases susceptibility through dual mechanisms (i.e., directly upregulating CsLOB1 expression and promoting cell wall loosening to facilitate bacterial entry). This study highlighted the importance of targeting transcription factors that regulate susceptibility genes as an alternative to modifying them. In rice, Oliva et al. developed a particularly effective strategy against bacterial blight by editing the promoter elements of OsSWEET genes targeted by bacterial TAL effectors41. Field trials in the Philippines confirmed that these edited plants demonstrated lasting resistance against multiple strains of Xanthomonas oryzae pv. oryzae without any yield penalties.
Viral Disease Resistance
CRISPR-Cas9 has enabled innovative approaches to managing viral diseases in crops. Tripathi et al. addressed a unique challenge in banana breeding by targeting integrated endogenous banana streak virus (eBSV) sequences found in the banana’s B genome42. These viral sequences can activate spontaneously under stress conditions, limiting the crossbreeding potential with B-genome bananas. The CRISPR-Cas9 system effectively inactivated these integrated viral sequences, with about 75% of edited plants remaining virus-free even in water-stress conditions. This breakthrough overcame a significant breeding obstacle that had hindered banana improvement for decades.
For cassava, Gomez et al. (2019) edited two eIF4E isoforms (nCBP-1 and nCBP-2) that viruses typically co-opt for replication43. These edited cassava plants displayed reduced severity of cassava brown streak disease symptoms and lower virus accumulation. Field trials in Uganda demonstrated a significantly lower disease incidence under natural infection conditions, highlighting the potential of targeting host factors essential for viral replication as an effective strategy for developing virus-resistant varieties in vegetatively propagated crop crops.
Field Validation and Translation
Several disease resistance studies advanced beyond greenhouse testing to field validation, particularly Oliva et al. for bacterial blight resistance in rice and Gomez et al. for viral resistance in cassava41,44. These field trials confirmed that CRISPR-mediated disease resistance remains effective under natural conditions, marking a crucial step toward practical deployment. Numerous studies indicate that genetic modifications do not result in crop yield loss or unexpected changes in plant traits.
While CRISPR-Cas9 biotechnology targets globally important crops, the studies reviewed showed that there are geographic variations in their focus and application. For instance, rice field trials were commonly conducted in Asia, particularly in India, Philippines, and India, where rice is a staple crop and its infrastructure is well-developed41. On the other hand, cassava and banana field trials were commonly conducted in Africa, particularly in Uganda and Kenya42,44. The patterns suggested from the studies suggest that CRISPR interventions are heavily influenced by regional crop preferences and cause disparities in infrastructure and governance regulations.
Nutritional Enhancement
Enhancement of Health-Promoting Compounds
CRISPR-Cas9 has effectively increased levels of bioactive compounds with potential health benefits in food crops. Nonaka et al. demonstrated a practical approach to enhance γ-aminobutyric acid (GABA) content in tomato fruits45. This non-protein amino acid provides several health benefits, including anti-hypertensive effects. By creating targeted mutations in the C-terminal autoinhibitory domains of glutamate decarboxylase genes (SlGAD2 and SlGAD3), the researchers achieved up to a 15-fold increase in GABA accumulation in ripe tomato fruits compared to wild-type controls. Remarkably, these significant metabolic changes occurred without impacting fruit development, plant morphology, or other quality traits.
Oil Quality Improvement
Modifying the fatty acid composition in oilseed crops is another important nutritional enhancement objective. Do et al. utilized CRISPR-Cas9 to simultaneously target both homologous copies of the fatty acid desaturase genes (FAD2-1A and FAD2-1B) in soybeans46. They achieved highly efficient mutation rates (>90%) by employing dual guide RNAs. They dramatically altered the oil composition—raising the oleic acid content from approximately 20% to 80% while reducing linoleic acid from about 50% to 4%. The resulting high-oleic oil profile offers improved oxidative stability, extended shelf life, and enhanced health benefits compared to conventional soybean oil. This study demonstrated that CRISPR-Cas9 effectively modifies polyploid crops with multiple gene copies.
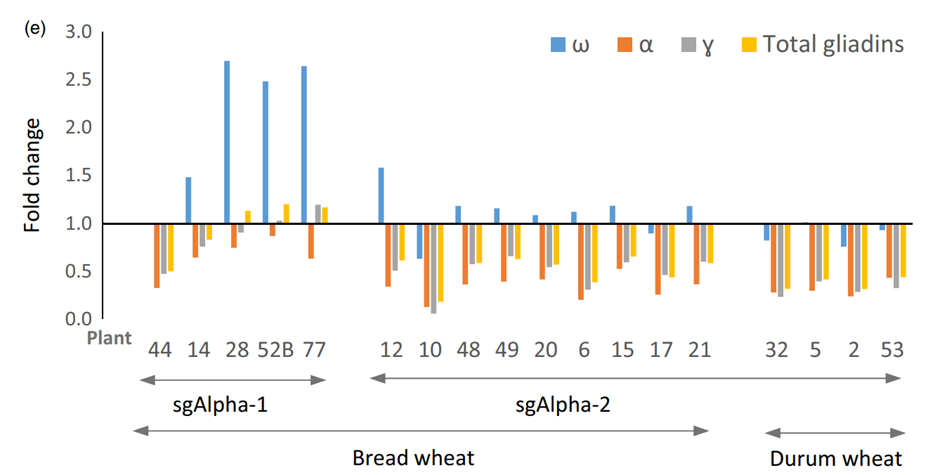
CRISPR-Cas9 has also significantly reduced allergens and anti-nutritional factors in staple foods. Sánchez-León et al. addressed a primary health concern by targeting multiple α-gliadin genes in bread wheat to decrease gluten content47. As shown in Figure 3, using CRISPR-Cas9, they successfully mutated up to 35 of the approximately 45 α-gliadin gene copies, reducing gluten content by up to 85%. Immunoreactivity assays utilizing antibodies from celiac disease patients confirmed a significant decrease in immunoreactivity in the modified wheat while maintaining the functional properties necessary for bread-making.
Mineral Biofortification
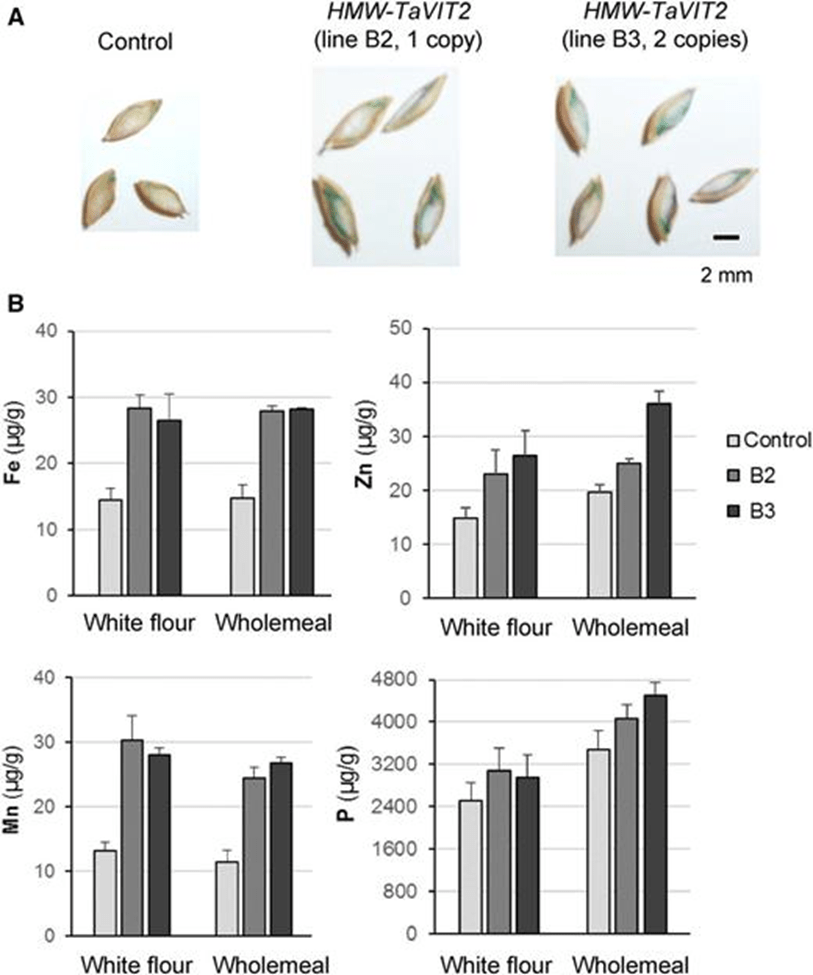
Micronutrient deficiencies, particularly iron deficiency, affect millions of people worldwide. Connorton et al. addressed this challenge by characterizing and manipulating the wheat vacuolar iron transporter TaVIT248. Their research indicated that TaVIT2 is involved in the transport of iron and manganese and that overexpressing this gene specifically in the endosperm significantly boosted the iron concentration in white flour by approximately two-fold compared to control lines. Notably, the biofortified wheat exhibited no yield penalty or impact on grain size, and the iron that accumulated was in a form readily bioavailable for human consumption. This can be seen from figure 4, where all iron, zinc, manganese, and phosphorus content increased for genetically modified crops, both B2 and B3, compared to control groups.
Vitamin Enhancement
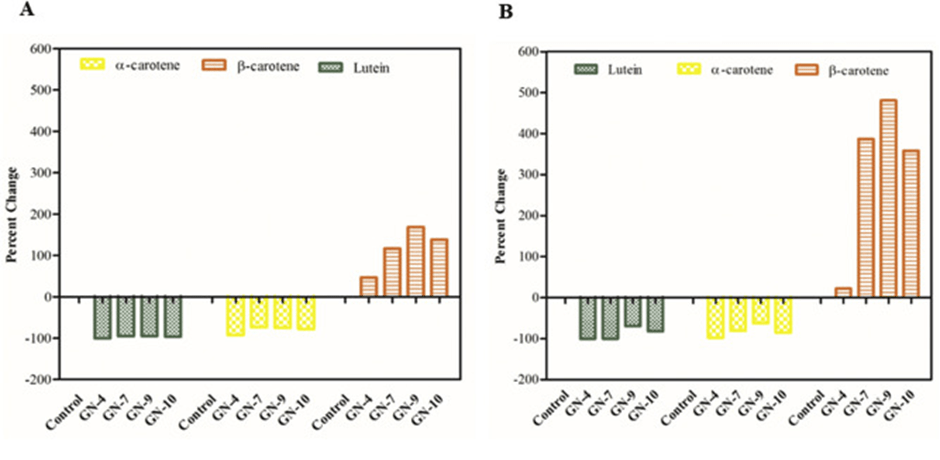
Vitamin A deficiency is a significant global health issue, especially in areas where bananas are a dietary staple. Kaur et al. utilized CRISPR-Cas9 to knock out the lycopene epsilon-cyclase gene (LCYε) in bananas, redirecting metabolic flow in the carotenoid biosynthetic pathway toward β-carotene (provitamin A) production49. As shown in Figure 5, this targeted change resulted in a six-fold increase in β-carotene content in edited banana fruits without impacting other important agronomic or fruit quality traits.
Climate Resilience Development
Salt Tolerance Enhancement
Soil salinity affects approximately 20% of irrigated agricultural land worldwide, making salt tolerance a crucial trait for maintaining crop productivity in many regions. Zhang et al. successfully employed CRISPR-Cas9 to knock out OsRR22, a negative regulator in the cytokinin signaling pathway in rice50. The edited rice plants showed better salt stress tolerance than the wild-type plants. Under high salinity conditions, the rice demonstrated higher survival rates with fewer damage symptoms by maintaining lower Na+/K+ ratios in shoots during salt stress, a vital indicator of enhanced salt tolerance. Notably, the knockout plants exhibited normal growth and development with no noticeable changes in key agronomic traits under standard growing conditions, suggesting that targeting negative regulators of stress responses provides a viable approach for crop improvement. The panel (a) of Figure 6 demonstrates the enhanced growth. The panel (b) of Figure 6 supports that CRISPR-edited crops show stronger resistance to saline environments , as the weights of CRISPR-edited rice plants (lighter and darker grey bars) are higher than those of wild types (white bars).
Liu et al. characterized the molecular mechanism of GmSALT3-mediated salt tolerance in soybean51. Although not directly employing CRISPR-Cas9 (thus creating a target for future editing), this study revealed that GmSALT3 confers salt tolerance primarily through chloride (Cl-) exclusion from leaves before sodium (Na+) exclusion. Field trials demonstrated significant differences in seed yields between near-isogenic lines with functional versus non-functional GmSALT3 under saline conditions. Interestingly, the gene did not enhance early seedling vigor under salinity, indicating its effects primarily manifest at later growth stages.

Drought Tolerance Improvement
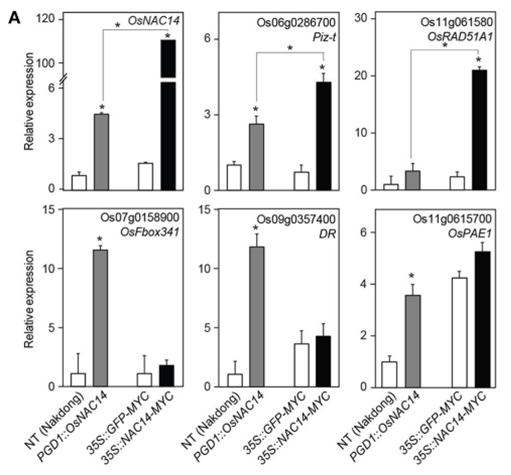
Drought represents one of the most significant climate-related threats to global crop production. Shim et al. demonstrated that the overexpression of the NAC transcription factor OsNAC14 in rice significantly enhanced drought tolerance52. Through comprehensive transcriptome analysis and chromatin immunoprecipitation (ChIP) experiments, they identified the direct target genes of OsNAC14, including those involved in stress response, ROS detoxification, and hormone signaling. It maintained higher relative water content and exhibited increased survival rates under severe drought conditions. Enhanced root development was also observed, improving water uptake in water-limited conditions. This study established OsNAC14 as a master regulator in a gene network controlling drought response, providing a valuable target for CRISPR-mediated enhancement.
In a study from Shim et al, expression of OsNAC14 in crops, especially in rice, revealed significant effects of drought tolerance and yield-related traits with stress conditions. OsNAC14-expressed plants showed an increase in panicles per hill by 18.62% compared to non-transgenic control under drought stress. Grain filling rate got improved as well where it increased from 12.25 to 36.64%, which indicates greater reproductive result. In addition, as shown in Figure 7, re-watered plants showed from 83 to 92% survival rate whereas NT plants showed 12%. Overall, this study revealed the effect of OsNAC14 as genetically targeted crops showed resilience to drought.
Shi et al. employed an innovative approach using CRISPR-Cas9 for targeted promoter replacement rather than gene knockout in maize53. By replacing the native promoter of ARGOS8 with the moderately constitutive GOS2 promoter, they created variants with enhanced expression under drought conditions. Field trials demonstrated that the edited plants showed approximately a 5% yield increase in water-limited environments without any yield penalty under well-watered conditions. The CRISPR-modified plants exhibited reduced inhibition of plant and ear growth under stress and decreased sensitivity to ethylene.
Cold Tolerance Development
Low-temperature stress significantly limits the geographical distribution and growing seasons of many crops, especially those of tropical or subtropical origin. Zhang et al.identified OsGATA16, a GATA transcription factor, as a positive regulator of cold tolerance in rice seedlings54. They demonstrated that OsGATA16 enhances cold tolerance by directly binding to the promoter of OsWRKY45-1 (a negative regulator of cold tolerance) and repressing its expression. Transgenic rice lines overexpressing OsGATA16 displayed significantly improved cold tolerance, while RNAi-mediated knockdown lines showed increased cold sensitivity. Under cold stress, the overexpressing lines maintained higher survival rates, chlorophyll content, and lower reactive oxygen species (ROS) accumulation than wild-type plants. This study uncovered a novel transcriptional regulatory module (OsGATA16-OsWRKY45-1) that could be targeted to enhance cold tolerance in rice and potentially other crops.
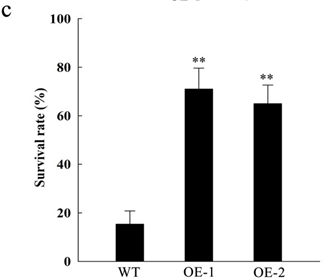
In Figure 8, It can be seen that WT shows survival rate about 17% whereas OE-1 and OE-2 shows about 70% and 67%, respectively. This reveals that genetic modification made it possible for crops to fight against cold conditions. The difference between WT and OE-1 or OE-2 is about 4 times, which OsGATA16 had a beneficial effect on price crops54.
Field Validation and Practical Application
In several instances, the shift to field validation was an auspicious aspect of climate resilience studies. The performance of CRISPR-edited ARGOS8 variants under drought stress, along with the validation of OsNAC14 overexpression lines in field trials, illustrated that improvements observed in the laboratory can be viable in real-world conditions. Additionally, the OsRR22 knockout rice demonstrated stable yields under normal conditions.
Yield and Quality Improvement
De Novo Domestication of Wild Species
Zsögön et al. pioneered an innovative method using CRISPR-Cas9 to simultaneously edit six domestication-related genes in wild tomato (Solanum pimpinellifolium)55. The researchers targeted genes controlling plant architecture (SELF-PRUNING and SELF-PRUNING 5G), fruit size (CLAVATA3 and WUSCHEL), and fruit shape (OVATE and FASCIATED). This multiplex editing transformed wild tomatoes into plants with domesticated characteristics while preserving advantageous traits from the wild germplasm.
The edited plants exhibited remarkable improvements across several agronomic traits: three times larger fruits, a tenfold increase in fruit number, altered fruit shape for easier harvesting, five times higher lycopene content than cultivated varieties, and a compact determinate growth structure suitable for modern agriculture.
Implications for Crop Improvement
This de novo domestication approach establishes a new paradigm for crop improvement that is particularly valuable in addressing climate change challenges. The strategy offers significant advantages over traditional breeding: dramatically accelerated timelines, the precise introduction of traits without linkage drag, the ability to work with sexually incompatible wild species, and the preservation of valuable stress resistance traits from wild germplasm.
The success of multiplex genome editing in wild tomatoes demonstrates the potential for applying strategies similar to those of other crops, especially orphan crops and those with narrow genetic bases. The ability to simultaneously modify multiple yield-related genes represents a powerful tool for crop improvement previously unattainable with conventional breeding or earlier genetic engineering techniques.
Among the 20 studies in Table 1, approximately 35% (7 out of 20 studies) included field trials, either as standalone or in combination. The remaining, 65% of the studies, were conducted in controlled environments like greenhouses or labs. The imbalance between the types of studies, either fully controlled or not, demonstrates that CRISPR-Cas9 applications still require field-level validation.
Biosafety and Risk Concerns
While CRISPR-Cas9 is a precise genome-editing biotechnology, safety concerns—including off-target effects at untargeted loci of genes and ecological risks—still remain. Such mutations can lead to unwanted or unpredictable phenotypic traits. The potential for gene flow to the environment, where genome-edited organisms are not welcome, raises environmental concerns. These issues call for further investigation into how genome-edited crops should be managed in environmental settings. There are concerns regarding the ethics of using CRISPR-Cas9 technology. There would be inequality in accessing technology and unwanted mutation. First, not only the CRISPR-Cas9 product but also other genetically modified crops are expensive. Use of CRISPR-Cas9 is also expected to require a large amount of income from farmers to access it, which also means that those who receive low payment can hardly use this technology56. This eventually leads to increasing class differences between high- and low-income farmers. Second, Mark et al. revealed that with errors caused by CRISPR-Cas9, such as off-target effects, this potentially causes unwanted consequences to the human body once mutated crops are consumed. Even though this technology is highly precise, it is still possible to affect organisms in negative ways, either directly or indirectly57.
Regional Applications in Developing Countries
Severity of Crop Challenges by Region
Table 2: Agronomic and Environmental Challenges in Developing Regions This comprehensive overview synthesizes critical agricultural challenges across developing regions, quantifying the significant impact of pathogenic infections, nutrient deficiencies, and climate-related stressors on staple crop production. The tabulated data illustrates the complex nature of agricultural constraints, demonstrating yield reductions, pathogen diversity, and soil nutrient limitations that threaten food security in vulnerable agricultural communities, ecosystems.
| Region | Crop | Pathogen/Challenge | Impact | Citation |
| Tanzania | Cassava | Cassava Brown Streak Disease (CBSD) | 30% to complete crop failure | Hillocks et al.58 |
| East and Central Africa | Cassava | Cassava Mosaic Disease (CMD) | 63% yield reduction, 43 million metric tons lost | Legg& Fauquet; Owor et al.59,60 |
| Nigeria | Banana/Plantain | Banana Bunchy Top Virus (BBTV) | Significant production threat | Adegbola et al.61 |
| African agroecological zones | Cassava | Xanthomonas axonopodis pv. manihotis | 18 distinct pathotypes identified | Wydra et al.62 |
| Indian subcontinent | Agricultural Soils | Zinc Deficiency | 49% of soils affected | Cakmak; Stein et al.63,64 |
| Eastern India | Rice | Drought Stress | 24-84% yield reduction | Dar et al.65 |
Viral, bacterial, and fungal pathogens threaten food security across developing regions, particularly impacting staple crops essential for subsistence agriculture. Cassava (Manihot esculenta), a primary caloric source for approximately 800 million people in Africa, is vulnerable to various devastating pathogens. Cassava brown streak disease (CBSD) causes yield reductions ranging from 30% to total crop failure in Tanzania66. Cassava mosaic disease (CMD), caused by several species of geminivirus, results in annual production losses estimated at 43 million metric tons across East and Central Africa67. Quantitative assessments in Uganda show that CMD infection reduces root yields by 63% in susceptible cultivars, with similar reductions observed in vegetative parameters68. Musa species (banana and plantain) cultivation face similar pathogenic challenges. The banana bunchy top virus (BBTV), recently identified in Nigeria, poses a significant threat to banana production systems in West Africa69. The prevalence of diverse bacterial pathogens complicates disease management strategies, illustrated by the identification of 18 distinct pathotypes of Xanthomonas axonopodis pv—manihotis affecting cassava production across African agroecological zones70.
Despite sufficient caloric intake in many regions, micronutrient deficiencies remain a major nutritional challenge, especially in sub-Saharan Africa71. In the Indian subcontinent, zinc deficiency impacts approximately 49% of agricultural soils, contributing to widespread zinc deficiency in human populations, with an estimated prevalence of 26%72,73.
Abiotic stresses linked to climate variability further worsen crop production limitations. Rainfed rice ecosystems in Eastern India, covering over 30 million hectares, experience yield reductions of 24-84% during periods of drought74. The Indo-Gangetic plains, supporting a population of over 400 million, are increasingly vulnerable to climate variability, negatively affecting both yield parameters and the nutritional composition of rice (Oryza sativa), the primary staple crop in the region75.
Enhancing Disease Resistance
CRISPR-Cas9 genome editing technology provides precise molecular interventions to address pathogen susceptibility in crops grown in developing regions. Oliva et al. demonstrated the successful field implementation of CRISPR-edited rice exhibiting broad-spectrum resistance to bacterial blight in the Philippines41. The researchers targeted promoter elements of susceptibility genes (OsSWEET), which are typically exploited by transcription activator-like (TAL) effectors from Xanthomonas oryzae pv. oryzae. Modifying these promoter elements hindered pathogen-induced activation of these genes, thereby conferring resistance to multiple bacterial strains. Field trials confirmed the durability of resistance without yield penalties, marking one of the initial successful field implementations of CRISPR-edited germplasm in the agricultural context of a developing country. In a comprehensive review of applications in Africa, Tripathi et al. documented ongoing research utilizing CRISPR-Cas9 to develop resistance against various viral diseases affecting staple crops, including cassava, banana, and sweet potato, which are crucial for food security across diverse African agroecological zones76.
Nutritional Enhancement
The application of CRISPR-Cas9 technology for nutritional biofortification offers a promising solution to address micronutrient deficiencies common in developing regions. Dong et al. reported the successful development of carotenoid-enriched rice through CRISPR-Cas9-mediated targeted gene insertion77. This research showed effective accumulation of β-carotene (provitamin A) in the rice endosperm, with the trait demonstrating stable inheritance across multiple generations. In addition, Endo et al. conducted the trials with targeting Osor gene in rice that resulted in an increase in β-carotene accumulation. This can be seen from the pronounced orange pigment of the tissues78. Establishing stable provitamin A biosynthesis in rice endosperm has significant implications for combating vitamin A deficiency, which affects an estimated 190 million preschool children globally, particularly in developing areas. Tripathi et al. identified several key targets for biofortification, including increasing provitamin A in cooking bananas, enhancing iron and zinc content in cereals, and improving protein quality in root crops76. They stressed that these nutritional enhancements could serve as sustainable solutions to combat hidden hunger, especially in rural communities that lack access to diverse food sources.
Climate Resilience Development
Santosh Kumar et al. reported the successful editing of the Drought and Salt Tolerance (OsDST) gene in MTU1010, an elite indica rice cultivar widely grown in rainfed ecosystems in India79. The researchers utilized CRISPR-Cas9 technology to target this negative regulator of stress tolerance, resulting in the creation of homozygous mutant lines that demonstrate improved resilience to drought and salinity stresses. Physiological analyses of the edited lines revealed significant enhancements in stress tolerance parameters, including better maintenance of relative water content, reduced membrane damage, elevated proline accumulation, and preservation of chlorophyll under stress conditions. The OsDST gene encodes a zinc finger transcription factor negatively regulating stress-responsive gene expression. By inactivating this gene through precise CRISPR-Cas9 editing, the researchers could upregulate stress-responsive genes, leading to increased tolerance without any apparent adverse effects on agronomic performance under non-stress conditions.
Implementation Challenges and Progress
The application of CRISPR-Cas9 technology in developing countries continues to face numerous challenges. A number of nations lack clear and consistent guidelines for gene-edited crops, which contributes to uncertainty and hindering the adoption of this technology80. In Africa, inadequate biosafety facilities and ineffective regulatory systems complicate the safe implementation of CRISPR-Cas9 techniques81. Furthermore, most studies are conducted in laboratory or greenhouse settings, with few being evaluated under actual field conditions.
Nonetheless, there are also notable advancements. Field trials in the Philippines have shown that CRISPR-edited rice exhibits resistance to bacterial blight without affecting yield41. CRISPR-based biotechnology efforts have been proven with banana, cassava, and rice across Asia and Africa82,76,81. Additionally, establishing regional centers of excellence for genome editing has promoted the development of local expertise in developing countries76.
However, due to the high cost of infrastructure, the need for skilled personnel, regulatory uncertainty, and unequal access to CRISPR biotechnology, low-resource regions—particularly in parts of Africa and Asia—face significant challenges in scaling genome-editing applications. Many African countries, including Kenya, Nigeria, Ghana, and Uganda, lack fully developed biosafety approval frameworks and necessary laboratory infrastructure, limiting the scalability of CRISPR-Cas9 technologies83. Smallholder farmers in these regions also face a shortage of local expertise, making them reliant on international research centers for genome-editing innovation76.
Conclusions and Implications
This review indicates that CRISPR-Cas9 research in agriculture primarily targets disease resistance, nutritional enhancement, and climate resilience. In terms of disease resistance, the technology has been utilized to target susceptibility genes and modify regulatory elements to combat fungal infections like powdery mildew in wheat37, bacterial pathogens such as Xanthomonas oryzae in rice41, and even viral threats like the endogenous banana streak virus42. Efforts in nutritional enhancement have led to significant advances, including a 15-fold increase in GABA levels in tomatoes45, an 85% reduction in gluten immunoreactivity in wheat47, and a 6-fold increase in provitamin A content in bananas49. Regarding climate resilience, CRISPR modifications have increased salt tolerance in rice through OsRR22 knockout50, boosted drought resistance in maize via promoter replacement in ARGOS833, and improved cold tolerance in rice through transcription factor regulation54. Even with its demonstrated advantages, CRISPR-Cas9 encounters limited efficiency in some crops, and uncertain biosafety and approved systems, especially in developing countries. Notably, several studies from developing countries have been emphasized—such as field trials of CRISPR-edited rice in the Philippines and virus-resistant cassava in Uganda—demonstrating the practical potential of these innovations in regions facing severe food security challenges.
These findings emphasize the need for clear, science-based regulatory frameworks to guide using CRISPR-Cas9 in agriculture, particularly in developing nations where food insecurity is most pronounced. The review suggests that although many developing countries face regulatory and infrastructure obstacles80,81, there are encouraging examples of progress. For instance, successful field trials of CRISPR-edited crops in the Philippines have shown that these technologies can be effectively implemented even in resource-limited environments. To fully leverage CRISPR-Cas9’s potential in addressing global hunger, policies should differentiate between traditional transgenic methods (i.e., GMOs) and precise genome edits (i.e., CRISPR-Cas9) that can occur naturally or through conventional breeding, thereby streamlining approval processes. Furthermore, promoting international partnerships and regional centers of excellence can help develop local expertise and facilitate technology transfer to smallholder farmers in developing regions84. The regional disparities in CRISPR-Cas9 adoption underscore the need for regionally adapted governance, regulatory and infrastructure.
The findings of this review present valuable implications for multiple stakeholders, including farmers, plant breeders, policymakers, and biotechnology firms. Farmers can benefit from CRISPR-edited crops with improved yield of harvest that is disease resistance and climate resistance. This leads to reduced dependency on chemical fertilizers and pesticides, ensuring the safety of the crops. CRISPR-Cas9 allows plant breeders to precisely conduct genome editing. This leads to various crop development with targeted traits in fewer breeding cycles. In order to accelerate the safe implementation of CRISPR biotechnology, policymakers need to establish a regulatory framework that differentiates between CRISPR-edited crops and GMOs. Also, they develop simplified, scientifically grounded approval systems to enhance real-world adoption in food-insecure regions. Lastly, biotechnology firms can establish commercial seed pipelines in collaboration with public institutions, ensuring that CRISPR-edited crops are effectively regulated and aligned with public agricultural needs.
While the Philippines has demonstrated success in field trials of CRISPR-edited rice for bacterial blight resistance41, its domestic research infrastructure remains relatively underdeveloped. This lack of technical capacity may be hindered by uncertain regulatory environments, leading to unequal access to genome-editing technologies. To address these barriers, CRISPR infrastructure should be developed alongside regionally tailored biosafety regulations. In the initial stages, international partnerships can help bridge capacity gaps until independent, locally managed systems are established.
Through interdisciplinary collaboration, CRISPR-Cas9 has the potential to enhance crop performance significantly and contribute to alleviating global hunger in developing nations.
Limitations and Future Directions
This review has several limitations. First, the analysis primarily relies on peer-reviewed studies that report successful applications of CRISPR-Cas9 in crops. This may introduce publication bias by underrepresenting studies with negative or inconclusive results. Second, while many promising applications are identified across various crop species, most studies were conducted in controlled laboratory or greenhouse settings. Only a few of these studies conducted field trials. Consequently, the actual effectiveness of CRISPR-Cas9 in different environmental settings and agricultural methods remains unclear.
Additionally, this review follows a narrative approach, allowing for a broad synthesis of existing research and presenting limitations. Unlike systematic reviews incorporating statistical analysis, this approach does not provide quantitative measures such as effect sizes or statistical significance. This makes it difficult to compare the effectiveness of different CRISPR-Cas9 applications directly. Furthermore, without standardized assessment tools like risk-of-bias scoring, the methodological quality of the studies cannot be objectively evaluated. This means that studies with varying levels of rigor may have been assigned equal weight in the analysis. Lastly, the review only includes studies published between 2014 and 2024, indicating that ongoing or unpublished research may have been overlooked.
Future systematic reviews should address quantitative analyses, such as meta-analyses, to evaluate the effectiveness of CRISPR-Cas9 across different crops. This approach would yield measurable insights into effect sizes, statistical significance, and variability among studies. Additionally, implementing structured risk-of-bias assessments would help ensure a more objective evaluation of study quality. Future reviews should also prioritize research that includes field trials, as most current studies are confined to controlled environments, which makes it challenging to assess real-world performance. Lastly, interdisciplinary reviews integrating genomic research, agricultural science, and socioeconomic factors would offer a broader perspective on CRISPR’s feasibility, adoption challenges, and economic impact, ensuring a more comprehensive assessment of its role in sustainable agriculture.
Due to plant diseases, climate change, scarce resources or abiotic factors, global food insecurity affects more than 800 million people in the world. Introduction of CRISPR-CAS9 was one of innovative findings that suggested great potential for dealing such problems throughout world such as rice blast in the southern United states that made economic loss around 1.1 billion dollars , and drought in sub-Saharan that made food shortage. While many critiques see drawbacks and limitations of CRISPR-CAS9 such as off-target and formation of mutation, this genetic engineering method can allow crops to endure such high temperature,severe drought or extreme cold weather. Resilience of crops is made with this technology, where it shows huge potential. With its possible ethical, technical and regulatory challenges, CRISPR-CAS9 is going through comprehensive assessment.
Crop types
This section is to show crop focused parts with how each plant got enhanced their genes with Crispr Cas9 technology. Even though the technology used is the same, the ability of each crop to improve is all different. In this section, the blended information will be organized.
- Rice: Genetic technology improved on resistance to blast and bacteria blight. This is due to editing on genes OsERF922 and OsSWEET. The tolerance that rice got enhanced are salt, drought, and cold, which is possible due to modification on OsRR22, OsNAC14, and OsGATA16. Lastly, knocking out LCYε enables nutritional improvement.
- Wheat: By modifying TaMLO and TaEDR1, wheat gains the ability to resist disease. Nutritional value also got improved by reducing gluten through knockout of alpha gliadin and increased iron content with TaVIT2.
- Maize: Drought resistance is done with the editing promoter of ARGOS8, which resulted in an increase in yield, about 5%.
- Soybean: With modification of FAD2 gene, it leads to nutritional quality improvement with its oil production with 80% oleic acid. In addition, salt tolerance was enhanced through GmSALT3.
- Banana: Edit of eBSV sequence leads to 75% virus-free plants. Moreover, nutritional improvement was done with LCYε knockout, which increased β-carotene sixfold.
- Tomato: Powdery Mildew resistance achieved with SlMLO1 gene got deleted. With editing of SlGAD2 and SIGAD3 gene, GABA content increased. In addition, wild tomato improved its limitation with multiple editing of six domestication genes. This resulted in a threefold increase in fruit size, tenfold increase in their number , and fivefold increase in lycopene content.
- Cassava: Viral resistance was enhanced by knocking out nCBP-1 and nCBP-2, reducing cassava brown streak disease severity.
- Citrus: Resistance of disease, citrus canker, was enabled with gene editing, specifically CsLOB1 promoter and suppressing the WRKY22 transcription factor.
References
- FAO, IFAD, UNICEF, WFP, & WHO. The state of food security and nutrition in the world 2020. Transforming food systems for affordable healthy diets. FAO (2020). https://doi.org/10.4060/ca9692en [↩] [↩]
- L. Nalley, F. Tsiboe, A. Durand-Morat, A. Shew, & G. Thoma. Economic and environmental impact of rice blast pathogen (Magnaporthe oryzae) alleviation in the United States. PLOS ONE. 11, e0167295 (2016). https://doi.org/10.1371/journal.pone.0167295 [↩] [↩]
- J. E. Cairns, J. Hellin, K. Sonder, J. L. Araus, J. F. MacRobert, C. Thierfelder, & B. M. Prasanna. Adapting maize production to climate change in sub-Saharan Africa. Food Security. 5, 345-360 (2013). https://doi.org/10.1007/s12571-013-0256-x [↩] [↩]
- S. Savary, L. Willocquet, S. J. Pethybridge, P. Esker, N. McRoberts, & A. Nelson. The global burden of pathogens and pests on major food crops. Nature Ecology & Evolution. 3, 430-439 (2019). https://doi.org/10.1038/s41559-018-0793-y [↩]
- H. E. Bouis, & R. M. Welch. Biofortification—A sustainable agricultural strategy for reducing micronutrient malnutrition in the global south. Crop Science. 50, S-20-S-32 (2010). https://doi.org/10.2135/cropsci2009.09.0531 [↩]
- G. A. Stevens, J. E. Bennett, Q. Hennocq, Y. Lu, L. M. De-Regil, L. Rogers, G. Danaei, G. Li, R. A. White, S. R. Flaxman, S. P. Oehrle, M. M. Finucane, R. Guerrero, Z. A. Bhutta, A. Then-Paulino, W. Fawzi, R. E. Black, & M. Ezzati. Trends and mortality effects of vitamin A deficiency in children in 138 low-income and middle-income countries between 1991 and 2013: A pooled analysis of population-based surveys. The Lancet Global Health. 3, e528-e536 (2015). https://doi.org/10.1016/S2214-109X(15)00039-X [↩]
- S. S. Myers, M. R. Smith, S. Guth, C. D. Golden, B. Vaitla, N. D. Mueller, A. D. Dangour, & P. Huybers. Climate change and global food systems: Potential impacts on food security and undernutrition. Annual Review of Public Health. 38, 259-277 (2017). https://doi.org/10.1146/annurev-publhealth-031816-044356 [↩]
- H. Zhu, C. Li, & C. Gao. Applications of CRISPR–Cas in agriculture and plant biotechnology. Nature Reviews Molecular Cell Biology. 21, 661-677 (2020). https://doi.org/10.1038/s41580-020-00288-9 [↩]
- S. Ahmad, N. Munawar, & A. Jamil. GMOs or non-GMOs? The CRISPR Conundrum. Frontiers in Plant Science. 14, 1232938 (2023). https://doi.org/10.3389/fpls.2023.1232938 [↩]
- Y. Zhang, K. Massel, I. D. Godwin, & C. Gao. Applications and potential of genome editing in crop improvement. Genome Biology. 19, 210 (2018). https://doi.org/10.1186/s13059-018-1586-y [↩]
- H. Zhu, C. Li, & C. Gao. Applications of CRISPR–Cas in agriculture and plant biotechnology. Nature Reviews Molecular Cell Biology. 21, 661-677 (2020). https://doi.org/10.1038/s41580-020-00288-9 [↩] [↩]
- K. Chen, Y. Wang, R. Zhang, H. Zhang, & C. Gao. CRISPR/Cas genome editing and precision plant breeding in agriculture. Annual Review of Plant Biology. 70, 667-697 (2019). https://doi.org/10.1146/annurev-arplant-050718-100049 [↩]
- J. Metje-Sprink, J. Menz, D. Modrzejewski, & T. Sprink. DNA-free genome editing: Past, present and future. Frontiers in Plant Science. 9, 1957 (2019). https://doi.org/10.3389/fpls.2018.01957 [↩]
- S. S.-e.-A. Zaidi, H. Vanderschuren, M. Qaim, M. M. Mahfouz, A. Kohli, S. Mansoor, & M. Tester. New plant breeding technologies for food security. Science. 363, 1390-1391 (2019). https://doi.org/10.1126/science.aav6316 [↩]
- Y. Wang, X. Cheng, Q. Shan, Y. Zhang, J. Liu, C. Gao, & J. L. Qiu. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nature Biotechnology. 32, 947-951 (2014). https://doi.org/10.1038/nbt.2969 [↩]
- F. Wang, C. Wang, P. Liu, C. Lei, W. Hao, Y. Gao, Y. G. Liu, & K. Zhao. Enhanced rice blast resistance by CRISPR/Cas9-targeted mutagenesis of the ERF transcription factor gene OsERF922. PLOS ONE. 11, e0154027 (2016). https://doi.org/10.1371/journal.pone.0154027 [↩]
- V. Nekrasov, C. Wang, J. Win, C. Lanz, D. Weigel, & S. Kamoun. Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Scientific Reports. 7, 482 (2017). https://doi.org/10.1038/s41598-017-00578-x [↩]
- Q. Long, M. Du, J. Long, Y. Xie, J. Zhang, L. Xu, Y. He, Q. Li, S. Chen, & X. Zou. Transcription factor WRKY22 regulates canker susceptibility in sweet orange (Citrus sinensis Osbeck) by enhancing cell enlargement and CsLOB1 expression. Horticulture Research. 8, 50 (2021). https://doi.org/10.1038/s41438-021-00486-2 [↩]
- R. Oliva, C. Ji, G. Atienza-Grande, J. C. Huguet-Tapia, A. Perez-Quintero, T. Li, J.-S. Eom, C. Li, H. Nguyen, B. Liu, F. Auguy, C. Sciallano, V. A. T. Luu, G. S. Dossa, S. Cunnac, S. M. Schmidt, I. H. Slamet-Loedin, C. Vera Cruz, B. Szurek, W. B. Frommer, F. F. White, & B. Yang. Broad-spectrum resistance to bacterial blight in rice using genome editing. Nature Biotechnology. 37, 1344-1350 (2019). https://doi.org/10.1038/s41587-019-0267-z [↩]
- J. N. Tripathi, V. O. Ntui, M. Ron, S. K. Muiruri, A. Britt, & L. Tripathi. CRISPR/Cas9 editing of endogenous banana streak virus in the B genome of Musa spp. overcomes a major challenge in banana breeding. Communications Biology. 2, 46 (2019). https://doi.org/10.1038/s42003-019-0288-7 [↩]
- M. A. Gomez, Z. D. Lin, T. Moll, R. D. Chauhan, L. Hayden, K. Renninger, G. Beyene, N. J. Taylor, J. C. Carrington, B. J. Staskawicz, & R. S. Bart. Simultaneous CRISPR/Cas9-mediated editing of cassava eIF4E isoforms nCBP-1 and nCBP-2 reduces cassava brown streak disease symptom severity and incidence. Plant Biotechnology Journal. 17, 421-434 (2019). https://doi.org/10.1111/pbi.12987 [↩]
- H. Jia, Y. Zhang, V. Orbović, J. Xu, F. F. White, J. B. Jones, & N. Wang. Genome editing of the disease susceptibility gene CsLOB1 in citrus confers resistance to citrus canker. Plant Biotechnology Journal. 15, 817-823 (2017). https://doi.org/10.1111/pbi.12677 [↩]
- Y. Zhang, Y. Bai, G. Wu, S. Zou, Y. Chen, C. Gao, & D. Tang. Simultaneous modification of three homoeologs of TaEDR1 by genome editing enhances powdery mildew resistance in wheat. The Plant Journal. 91, 714-724 (2017). https://doi.org/10.1111/tpj.13599 [↩]
- S. Nonaka, C. Arai, M. Takayama, C. Matsukura, & H. Ezura. Efficient increase of ɣ-aminobutyric acid (GABA) content in tomato fruits by targeted mutagenesis. Scientific Reports. 7, 7057 (2017). https://doi.org/10.1038/s41598-017-06400-y [↩]
- P. T. Do, C. X. Nguyen, H. T. Bui, L. T. N. Tran, G. Stacey, J. D. Gillman, Z. J. Zhang, G. Stacey, & H. T. Nguyen. Demonstration of highly efficient dual gRNA CRISPR/Cas9 editing of the homeologous GmFAD2-1A and GmFAD2-1B genes to yield a high oleic, low linoleic and α-linolenic acid phenotype in soybean. BMC Plant Biology. 19, 311 (2019). https://doi.org/10.1186/s12870-019-1906-8 [↩]
- S. Sánchez-León, J. Gil-Humanes, C. V. Ozuna, M. J. Giménez, C. Sousa, D. F. Voytas, & F. Barro. Low-gluten, nontransgenic wheat engineered with CRISPR/Cas9. Plant Biotechnology Journal. 16, 902-910 (2018). https://doi.org/10.1111/pbi.12837 [↩]
- J. M. Connorton, E. R. Jones, I. Rodríguez-Ramiro, S. Fairweather-Tait, C. Uauy, & J. Balk. Wheat vacuolar iron transporter TaVIT2 transports Fe and Mn and is effective for biofortification. Plant Physiology. 174, 2434-2444 (2017). https://doi.org/10.1104/pp.17.00672 [↩]
- N. Kaur, A. Alok, Shivani, P. Kumar, N. Kaur, P. Awasthi, S. Chaturvedi, P. Pandey, A. Pandey, A. K. Pandey, & S. Tiwari. CRISPR/Cas9 directed editing of lycopene epsilon-cyclase modulates metabolic flux for β-carotene biosynthesis in banana fruit. Metabolic Engineering. 59, 76-86 (2020). https://doi.org/10.1016/j.ymben.2020.01.008 [↩]
- A. Zhang, Y. Liu, F. Wang, T. Li, Z. Chen, D. Kong, J. Bi, F. Zhang, X. Luo, J. Wang, J. Tang, X. Yu, G. Liu, & L. Luo. Enhanced rice salinity tolerance via CRISPR/Cas9-targeted mutagenesis of the OsRR22 gene. Molecular Breeding. 39, 47 (2019). https://doi.org/10.1007/s11032-019-0954-y [↩]
- J. S. Shim, N. Oh, P. J. Chung, Y. S. Kim, Y. D. Choi, & J. K. Kim. Overexpression of OsNAC14 improves drought tolerance in rice. Frontiers in Plant Science. 9, 310 (2018). https://doi.org/10.3389/fpls.2018.00310 [↩]
- Y. Liu, L. Yu, Y. Qu, J. Chen, X. Liu, H. Hong, Z. Liu, R. Chang, M. Gilliham, L. Qiu, & R. Guan. GmSALT3, which confers improved soybean salt tolerance in the field, increases leaf Cl- exclusion prior to Na+ exclusion but does not improve early vigor under salinity. Frontiers in Plant Science. 7, 1485 (2016). https://doi.org/10.3389/fpls.2016.01485 [↩]
- H. Zhang, T. Wu, Z. Li, K. Huang, N. E. Kim, Z. Ma, S. W. Kwon, W. Jiang, & X. Du. OsGATA16, a GATA transcription factor, confers cold tolerance by repressing OsWRKY45-1 at the seedling stage in rice. Rice. 14(1), 42 (2021). https://doi.org/10.1186/s12284-021-00485-w [↩]
- J. Shi, H. Gao, H. Wang, H. R. Lafitte, R. L. Archibald, M. Yang, S. M. Hakimi, H. Mo, & J. E. Habben. ARGOS8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnology Journal. 15(2), 207-216 (2017). https://doi.org/10.1111/pbi.12603 [↩] [↩]
- A. Zsögön, T. Čermák, E. R. Naves, M. M. Notini, K. H. Edel, S. Weinl, L. Freschi, D. F. Voytas, J. Kudla, & L. E. P. Peres. De novo domestication of wild tomato using genome editing. Nature Biotechnology. 36(12), 1211-1216 (2018). https://doi.org/10.1038/nbt.4272 [↩]
- H. Jia, Y. Zhang, V. Orbović, J. Xu, F. F. White, J. B. Jones, & N. Wang. Genome editing of the disease susceptibility gene CsLOB1 in citrus confers resistance to citrus canker. Plant Biotechnology Journal. 15, 817-823 (2017). https://doi.org/10.1111/pbi.12677 [↩] [↩]
- V. Nekrasov, C. Wang, J. Win, C. Lanz, D. Weigel, & S. Kamoun. Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Scientific Reports. 7, 482 (2017). https://doi.org/10.1038/s41598-017-00578-x [↩] [↩]
- Y. Wang, X. Cheng, Q. Shan, Y. Zhang, J. Liu, C. Gao, & J. L. Qiu. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nature Biotechnology. 32, 947-951 (2014). https://doi.org/10.1038/nbt.2969 [↩] [↩]
- Y. Zhang, Y. Bai, G. Wu, S. Zou, Y. Chen, C. Gao, & D. Tang. Simultaneous modification of three homoeologs of TaEDR1 by genome editing enhances powdery mildew resistance in wheat. The Plant Journal. 91, 714-724 (2017). https://doi.org/10.1111/tpj.13599 [↩]
- F. Wang, C. Wang, P. Liu, C. Lei, W. Hao, Y. Gao, Y. G. Liu, & K. Zhao. Enhanced rice blast resistance by CRISPR/Cas9-targeted mutagenesis of the ERF transcription factor gene OsERF922. PLOS ONE. 11, e0154027 (2016). https://doi.org/10.1371/journal.pone.0154027 [↩] [↩]
- Q. Long, M. Du, J. Long, Y. Xie, J. Zhang, L. Xu, Y. He, Q. Li, S. Chen, & X. Zou. Transcription factor WRKY22 regulates canker susceptibility in sweet orange (Citrus sinensis Osbeck) by enhancing cell enlargement and CsLOB1 expression. Horticulture Research. 8, 50 (2021). https://doi.org/10.1038/s41438-021-00486-2 [↩]
- R. Oliva, C. Ji, G. Atienza-Grande, J. C. Huguet-Tapia, A. Perez-Quintero, T. Li, J.-S. Eom, C. Li, H. Nguyen, B. Liu, F. Auguy, C. Sciallano, V. A. T. Luu, G. S. Dossa, S. Cunnac, S. M. Schmidt, I. H. Slamet-Loedin, C. Vera Cruz, B. Szurek, W. B. Frommer, F. F. White, & B. Yang. Broad-spectrum resistance to bacterial blight in rice using genome editing. Nature Biotechnology. 37, 1344-1350 (2019). https://doi.org/10.1038/s41587-019-0267-z [↩] [↩] [↩] [↩] [↩] [↩] [↩]
- J. N. Tripathi, V. O. Ntui, M. Ron, S. K. Muiruri, A. Britt, & L. Tripathi. CRISPR/Cas9 editing of endogenous banana streak virus in the B genome of Musa spp. overcomes a major challenge in banana breeding. Communications Biology. 2, 46 (2019). https://doi.org/10.1038/s42003-019-0288-7 [↩] [↩] [↩]
- M. A. Gomez, Z. D. Lin, T. Moll, R. D. Chauhan, L. Hayden, K. Renninger, G. Beyene, N. J. Taylor, J. C. Carrington, B. J. Staskawicz, & R. S. Bart. Simultaneous CRISPR/Cas9-mediated editing of cassava eIF4E isoforms nCBP-1 and nCBP-2 reduces cassava brown streak disease symptom severity and incidence. Plant Biotechnology Journal. 17, 421-434 (2019). https://doi.org/10.1111/pbi.12987 [↩]
- M. A. Gomez, Z. D. Lin, T. Moll, R. D. Chauhan, L. Hayden, K. Renninger, G. Beyene, N. J. Taylor, J. C. Carrington, B. J. Staskawicz, & R. S. Bart. Simultaneous CRISPR/Cas9-mediated editing of cassava eIF4E isoforms nCBP-1 and nCBP-2 reduces cassava brown streak disease symptom severity and incidence. Plant Biotechnology Journal. 17, 421-434 (2019). https://doi.org/10.1111/pbi.12987 [↩] [↩]
- S. Nonaka, C. Arai, M. Takayama, C. Matsukura, & H. Ezura. Efficient increase of ɣ-aminobutyric acid (GABA) content in tomato fruits by targeted mutagenesis. Scientific Reports. 7, 7057 (2017). https://doi.org/10.1038/s41598-017-06400-y [↩] [↩]
- P. T. Do, C. X. Nguyen, H. T. Bui, L. T. N. Tran, G. Stacey, J. D. Gillman, Z. J. Zhang, G. Stacey, & H. T. Nguyen. Demonstration of highly efficient dual gRNA CRISPR/Cas9 editing of the homeologous GmFAD2-1A and GmFAD2-1B genes to yield a high oleic, low linoleic and α-linolenic acid phenotype in soybean. BMC Plant Biology. 19, 311 (2019). https://doi.org/10.1186/s12870-019-1906-8 [↩]
- S. Sánchez-León, J. Gil-Humanes, C. V. Ozuna, M. J. Giménez, C. Sousa, D. F. Voytas, & F. Barro. Low-gluten, nontransgenic wheat engineered with CRISPR/Cas9. Plant Biotechnology Journal. 16, 902-910 (2018). https://doi.org/10.1111/pbi.12837 [↩] [↩] [↩]
- J. M. Connorton, E. R. Jones, I. Rodríguez-Ramiro, S. Fairweather-Tait, C. Uauy, & J. Balk. Wheat vacuolar iron transporter TaVIT2 transports Fe and Mn and is effective for biofortification. Plant Physiology. 174, 2434-2444 (2017). https://doi.org/10.1104/pp.17.00672 [↩] [↩]
- N. Kaur, A. Alok, Shivani, P. Kumar, N. Kaur, P. Awasthi, S. Chaturvedi, P. Pandey, A. Pandey, A. K. Pandey, & S. Tiwari. CRISPR/Cas9 directed editing of lycopene epsilon-cyclase modulates metabolic flux for β-carotene biosynthesis in banana fruit. Metabolic Engineering. 59, 76-86 (2020). https://doi.org/10.1016/j.ymben.2020.01.008 [↩] [↩] [↩]
- A. Zhang, Y. Liu, F. Wang, T. Li, Z. Chen, D. Kong, J. Bi, F. Zhang, X. Luo, J. Wang, J. Tang, X. Yu, G. Liu, & L. Luo. Enhanced rice salinity tolerance via CRISPR/Cas9-targeted mutagenesis of the OsRR22 gene. Molecular Breeding. 39, 47 (2019). https://doi.org/10.1007/s11032-019-0954-y [↩] [↩] [↩]
- Y. Liu, L. Yu, Y. Qu, J. Chen, X. Liu, H. Hong, Z. Liu, R. Chang, M. Gilliham, L. Qiu, & R. Guan. GmSALT3, which confers improved soybean salt tolerance in the field, increases leaf Cl- exclusion prior to Na+ exclusion but does not improve early vigor under salinity. Frontiers in Plant Science. 7, 1485 (2016). https://doi.org/10.3389/fpls.2016.01485 [↩]
- J. S. Shim, N. Oh, P. J. Chung, Y. S. Kim, Y. D. Choi, & J. K. Kim. Overexpression of OsNAC14 improves drought tolerance in rice. Frontiers in Plant Science. 9, 310 (2018). https://doi.org/10.3389/fpls.2018.00310 [↩] [↩]
- J. Shi, H. Gao, H. Wang, H. R. Lafitte, R. L. Archibald, M. Yang, S. M. Hakimi, H. Mo, & J. E. Habben. ARGOS8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnology Journal. 15(2), 207-216 (2017). https://doi.org/10.1111/pbi.12603 [↩]
- H. Zhang, T. Wu, Z. Li, K. Huang, N. E. Kim, Z. Ma, S. W. Kwon, W. Jiang, & X. Du. OsGATA16, a GATA transcription factor, confers cold tolerance by repressing OsWRKY45-1 at the seedling stage in rice. Rice. 14(1), 42 (2021). https://doi.org/10.1186/s12284-021-00485-w [↩] [↩] [↩] [↩]
- \0A. Zsögön, T. Čermák, E. R. Naves, M. M. Notini, K. H. Edel, S. Weinl, L. Freschi, D. F. Voytas, J. Kudla, & L. E. P. Peres. De novo domestication of wild tomatoes using genome editing. Nature Biotechnology. 36(12), 1211-1216 (2018). https://doi.org/10.1038/nbt.4272 [↩]
- Subica, A. M. (2023). CRISPR in Public Health: The Health Equity Implications and Role of Community in Gene-Editing Research and Applications. American Journal of Public Health, 113(8), 874–882. https://doi.org/10.2105/ajph.2023.307315 [↩]
- Mark, Pieter, J., Lianne, Adinda De Schrijver, Ruud, Kleter, G. A., & Wilde, E. B. (2022). Occurrence and Nature of Off-Target Modifications by CRISPR-Cas Genome Editing in Plants. ACS Agricultural Science & Technology, 2(2), 192–201. https://doi.org/10.1021/acsagscitech.1c00270 [↩]
- R. J. Hillocks, M. D. Raya, K. Mtunda, & H. Kiozia. Effects of brown streak virus disease on yield and quality of cassava in Tanzania. Journal of Phytopathology. 149(7-8), 389-394 (2001). https://doi.org/10.1111/j.1439-0434.2001.tb03868.x [↩]
- J. P. Legg, & C. M. Fauquet. Cassava mosaic geminiviruses in Africa. Plant Molecular Biology. 56(4), 585-599 (2004). https://doi.org/10.1007/s11103-004-1651-7 [↩]
- B. Owor, J. P. Legg, G. Okao-Okuja, R. Obonyo, & M. W. Ogenga-Latigo. The effect of cassava mosaic geminiviruses on symptom severity, growth and root yield of a cassava mosaic virus disease-susceptible cultivar in Uganda. Annals of Applied Biology. 145(3), 331-337 (2004). https://doi.org/10.1111/j.1744-7348.2004.tb00390.x [↩]
- R. O. Adegbola, A. O. Ayodeji, A. C. Awosusi, G. I. Atiri, & P. L. Kumar. First report of banana bunchy top virus in banana and plantain (Musa spp.) in Nigeria. Plant Disease. 97(2), 290 (2013). https://doi.org/10.1094/PDIS-08-12-0745-PDN [↩]
- K. Wydra, V. Zinsou, V. Jorge, & V. Verdier. Identification of pathotypes of Xanthomonas axonopodis pv. manihotis in Africa and detection of quantitative trait loci and markers for resistance to bacterial blight of cassava. Phytopathology. 94(10), 1084-1093 (2004). https://doi.org/10.1094/PHYTO.2004.94.10.1084 [↩]
- I. Cakmak. Enrichment of fertilizers with zinc: An excellent investment for humanity and crop production in India. Journal of Trace Elements in Medicine and Biology. 23(4), 281-289 (2009). https://doi.org/10.1016/j.jtemb.2009.05.002 [↩]
- A. J. Stein, P. Nestel, J. V. Meenakshi, M. Qaim, H. P. S. Sachdev, & Z. A. Bhutta. Plant breeding to control zinc deficiency in India: How cost-effective is biofortification? Public Health Nutrition. 10(5), 492-501 (2007). https://doi.org/10.1017/S1368980007223857 [↩]
- M. H. Dar, C. H. Waza, S. G. Shukla, N. K. Chaya, S. Neeraj, J. S. Soumya, & U. S. Singh. Drought tolerant rice for ensuring food security in eastern India. Sustainability. 12(16), 6490 (2020). https://doi.org/10.3390/su12062214 [↩]
- R. J. Hillocks, M. D. Raya, K. Mtunda, & H. Kiozia. Effects of brown streak virus disease on yield and quality of cassava in Tanzania. Journal of Phytopathology. 149(7-8), 389-394 (2001). https://doi.org/10.1111/j.1439-0434.2001.tb03868.x [↩]
- J. P. Legg, & C. M. Fauquet. Cassava mosaic geminiviruses in Africa. Plant Molecular Biology. 56(4), 585-599 (2004). https://doi.org/10.1007/s11103-004-1651-7 [↩]
- B. Owor, J. P. Legg, G. Okao-Okuja, R. Obonyo, & M. W. Ogenga-Latigo. The effect of cassava mosaic geminiviruses on symptom severity, growth and root yield of a cassava mosaic virus disease-susceptible cultivar in Uganda. Annals of Applied Biology. 145(3), 331-337 (2004). https://doi.org/10.1111/j.1744-7348.2004.tb00390.x [↩]
- R. O. Adegbola, A. O. Ayodeji, A. C. Awosusi, G. I. Atiri, & P. L. Kumar. First report of banana bunchy top virus in banana and plantain (Musa spp.) in Nigeria. Plant Disease. 97(2), 290 (2013). https://doi.org/10.1094/PDIS-08-12-0745-PDN [↩]
- K. Wydra, V. Zinsou, V. Jorge, & V. Verdier. Identification of pathotypes of Xanthomonas axonopodis pv. manihotis in Africa and detection of quantitative trait loci and markers for resistance to bacterial blight of cassava. Phytopathology. 94(10), 1084-1093 (2004). https://doi.org/10.1094/PHYTO.2004.94.10.1084 [↩]
- A. W. de Valença, A. Bake, I. D. Brouwer, & K. E. Giller. Agronomic biofortification of crops to fight hidden hunger in sub-Saharan Africa. Global Food Security. 12, 8-14 (2017). https://doi.org/10.1016/j.gfs.2016.12.001 [↩]
- I. Cakmak. Enrichment of fertilizers with zinc: An excellent investment for humanity and crop production in India. Journal of Trace Elements in Medicine and Biology. 23(4), 281-289 (2009). https://doi.org/10.1016/j.jtemb.2009.05.002 [↩]
- A. J. Stein, P. Nestel, J. V. Meenakshi, M. Qaim, H. P. S. Sachdev, & Z. A. Bhutta. Plant breeding to control zinc deficiency in India: How cost-effective is biofortification? Public Health Nutrition. 10(5), 492-501 (2007). https://doi.org/10.1017/S1368980007223857 [↩]
- M. H. Dar, C. H. Waza, S. G. Shukla, N. K. Chaya, S. Neeraj, J. S. Soumya, & U. S. Singh. Drought tolerant rice for ensuring food security in eastern India. Sustainability. 12(16), 6490 (2020). https://doi.org/10.3390/su12062214 [↩]
- V. Pooniya, & Y. S. Shivay. Enrichment of basmati rice grain and straw with zinc and nitrogen through ferti-fortification and summer green manuring under Indo-Gangetic plains of India. Journal of Plant Nutrition. 36(1), 91-117 (2013). https://doi.org/10.1080/01904167.2012.733052 [↩]
- L. Tripathi, K. S. Dhugga, V. O. Ntui, S. Runo, E. D. Syombua, S. Muiruri, Z. Wen, & J. N. Tripathi. Genome editing for sustainable agriculture in Africa. Frontiers in Genome Editing. 4, 876697 (2022). https://doi.org/10.3389/fgeed.2022.876697 [↩] [↩] [↩] [↩] [↩]
- O. X. Dong, S. Yu, R. Jain, N. Zhang, P. Q. Duong, C. Butler, Y. Li, A. Lipzen, J. A. Martin, K. W. Barry, J. Schmutz, A. M. Tian, & P. C. Ronald. Marker-free carotenoid-enriched rice generated through targeted gene insertion using CRISPR-Cas9. Nature Communications. 11(1), 1178 (2020). https://doi.org/10.1038/s41467-020-14981-y [↩]
- Endo, A., Hiroaki Saika, Takemura, M., Misawa, N., & Toki, S. (2019). A novel approach to carotenoid accumulation in rice callus by mimicking the cauliflower Orange mutation via genome editing. Rice, 12(1). https://doi.org/10.1186/s12284-019-0345-3 [↩]
- V. V. Santosh Kumar, R. K. Verma, S. K. Yadav, P. Yadav, A. Watts, M. V. Rao, & V. Chinnusamy. CRISPR-Cas9 mediated genome editing of drought and salt tolerance (OsDST) genes in indica mega rice cultivar MTU1010. Physiology and Molecular Biology of Plants. 26(6), 1099-1110 (2020). https://doi.org/10.1007/s12298-020-00819-w [↩]
- C. Turnbull, M. Lillemo, & T. A. K. Hvoslef-Eide. Global regulation of genetically modified crops amid the gene edited crop boom–A review. Frontiers in Plant Science. 12, 630396 (2021). https://doi.org/10.3389/fpls.2021.630396 [↩] [↩]
- E. G. Kedisso, K. Maredia, J. Guenthner, & M. Koch. Commercialization of genetically modified crops in Africa: opportunities and challenges. African Journal of Biotechnology. 21(5), 188-197 (2022). https://doi.org/10.5897/AJB2021.17434 [↩] [↩] [↩]
- R. Oliva, C. Ji, G. Atienza-Grande, J. C. Huguet-Tapia, A. Perez-Quintero, T. Li, J.-S. Eom, C. Li, H. Nguyen, B. Liu, F. Auguy, C. Sciallano, V. A. T. Luu, G. S. Dossa, S. Cunnac, S. M. Schmidt, I. H. Slamet-Loedin, C. Vera Cruz, B. Szurek, W. B. Frommer, F. F. White, & B. Yang. Broad-spectrum resistance to bacterial blight in rice using genome editing. Nature Biotechnology. 37, 1344-1350 (2019) https://doi.org/10.1038/s41587-019-0267-z [↩]
- E. G. Kedisso, K. Maredia, J. Guenthner, & M. Koch. Commercialization of genetically modified crops in Africa: opportunities and challenges. African Journal of Biotechnology. 21(5), 188-197 (2022). https://doi.org/10.5897/AJB2021.17434 [↩]
- L. Tripathi, K. S. Dhugga, V. O. Ntui, S. Runo, E. D. Syombua, S. Muiruri, Z. Wen, & J. N. Tripathi. Genome editing for sustainable agriculture in Africa. Frontiers in Genome Editing. 4, 876697 (2022). https://doi.org/10.3389/fgeed.2022.876697 [↩]