Abstract
Chemical isotope patterns serve as molecular fingerprints in mass spectrometry for compound identification in analytical chemistry. Traditional discrete isotope pattern representations face significant computational limitations when integrated with modern machine learning frameworks, which require fixed-dimension numerical inputs for effective processing. This study develops and validates a Gaussian distribution modeling approach for converting discrete isotope patterns into continuous vector representations suitable for machine learning applications. Using 1,902 compounds acquired from PubChem database through stratified random sampling, we show that Gaussian modeling with optimized parameters (σ = 0.02 Da, 1000-dimensional vectors) achieves superior performance compared to traditional discrete methods across multiple validation metrics. Principal component analysis revealed that 85.2% of variance could be explained by the first 10 components, with PC1 correlating strongly with molecular weight (r = 0.89) and PC3 with structural complexity (r = 0.68). K-means clustering identified eight distinct chemical families with excellent separation (silhouette score = 0.742 vs 0.523 for discrete methods). Performance comparisons showed significant improvements in pattern similarity correlation (Gaussian: r = 0.996 ± 0.004 vs Discrete: r = 0.78 ± 0.07), clustering stability (94.3% vs 73.2%), and computational efficiency (0.067 vs 1.24 seconds per compound). This comparative analysis shows that Gaussian modeling provides a strong basis for next-generation chemical informatics platforms while preserving chemical
information with >99.5% fidelity.
Keywords: isotope patterns, Gaussian distribution, chemical informatics, machine learning, mass spectrometry, molecular fingerprints, principal component analysis, discrete pattern comparison
Introduction
Chemical isotope patterns are among the most information-rich characteristics of molecular species in analytical chemistry, particularly in high-resolution mass spectrometry applications, and these patterns act as unique molecular fingerprints that allow precise compound identification and structure determination in complex chemical mixtures1. These patterns arise from the statistical distribution of naturally occurring isotopes within molecular structures, creating distinctive spectral signatures that provide important information beyond simple molecular weight determination, including discrimination between isobaric compounds, validation of molecular formulas, and assessment of sample purity2. Isotope patterns are based on multinomial probability distributions, where each isotope’s natural abundance contributes to the overall pattern according to well-established combinatorial principles3.
Literature Review
Traditional Isotope Pattern Analysis Methods
Historical approaches to isotope pattern analysis have relied primarily on discrete peak-based representations that accurately capture the physics of mass spectrometry but present significant computational challenges for modern applications4. Early computational methods for isotope pattern calculation, such as the polynomial method developed by Yergey5 and the pruning algorithms proposed by Rockwood et al.6, focused on accurate prediction but did not address integration with machine learning.
Recent advances in isotope pattern deconvolution have explored several mathematical approaches including Fourier transform methods7, maximum likelihood estimation8, and Bayesian inference techniques9. However, these methods still use discrete representations that require specialized preprocessing before integration with standard machine learning algorithms without specialized preprocessing10.
Machine Learning Applications in Mass Spectrometry
The integration of machine learning with mass spectrometry data has shown promising results across various applications including compound identification11, metabolomics analysis12, and spectral library matching13. Deep learning approaches have been particularly successful for spectral prediction14 and structural elucidation15, but these methods typically bypass isotope pattern information due to representation challenges. Continuous vector representations of molecular data have proven essential for effective machine learning integration, as demonstrated by molecular fingerprinting methods16 and chemical space visualization techniques17. However, existing approaches for isotope pattern vectorization lack systematic validation and theoretical foundation18.
Gaussian Kernel Methods in Chemical Informatics
Gaussian kernel density estimation has been successfully applied to various chemical informatics problems including molecular similarity assessment19 and spectral data processing20. While Gaussian modeling approaches have been utilized in mass spectrometry for peak fitting and noise reduction21, systematic application to isotope pattern vectorization has not been comprehensively studied or validated against traditional discrete methods.
Methodology
Research Design and Ten-Stage Pipeline
This study employs a comprehensive computational chemistry approach combining large-scale data analysis, mathematical modeling, and statistical validation techniques through a systematic ten-stage pipeline methodology designed to ensure reproducibility and statistical rigor:
- Stage 1: Database selection and access protocol establishment
- Stage 2: Stratified sampling design implementation
- Stage 3: Compound data retrieval and molecular descriptor extraction
- Stage 4: Quality control filtering and validation procedures
- Stage 5: Theoretical isotope pattern calculation using multinomial expansion
- Stage 6: Gaussian distribution modeling parameter optimization
- Stage 7: Vector representation generation and standardization
- Stage 8: Principal component analysis and dimensionality reduction
- Stage 9: Clustering analysis and chemical family identification
- Stage 10: Performance validation and cross-verification procedures
Data Collection and Sample Selection
Chemical compound data was acquired from the PubChem database using the PUG REST API with comprehensive error handling and rate limiting protocols22. Stratified random sampling was employed across four molecular weight categories to ensure representative coverage of chemical space diversity: small molecules (CID 1-10,000, MW < 200 Da), medium molecules (CID 10,001-100,000, MW 200-500 Da), large molecules (CID 100,001-500,000, MW 500-1000 Da), and very large molecules (CID 500,001-1,000,000, MW 1000-2000 Da), with 500 compounds targeted from each stratum.
The molecular weight constraint of 2000 Da was established to focus on small-to-medium sized molecules commonly encountered in pharmaceutical and metabolomics applications, where isotope pattern analysis is most critical for accurate compound identification. This range encompasses the vast majority of bioactive compounds while maintaining computational tractability for high-throughput analysis.
For each compound, comprehensive molecular descriptors were retrieved including molecular formula (required for isotope calculations), molecular weight and exact mass, IUPAC systematic name, canonical and isomeric SMILES notation, hydrogen bond donor and acceptor counts, topological polar surface area, octanol-water partition coefficient, rotatable bond count, heavy atom count, and molecular complexity score.
Training/Testing Split: The dataset was divided into training (80%, n=1,522) and testing (20%, n=380) sets using stratified sampling to maintain representative distribution across molecular weight categories.
Quality Control and Filtering
Comprehensive quality control procedures were implemented to ensure dataset integrity through systematic filtering based on: (1) presence of valid molecular formula with standard elements only, (2) molecular weight constraints (12-2000 Da) to exclude fragments and polymers, (3) heavy atom count validation (1-100 atoms) to ensure reasonable molecular size, (4) data completeness assessment requiring all essential descriptors, and (5) duplicate removal based on identical molecular formulas. From an initial sample of 2,000 compounds, this rigorous process yielded 1,902 unique, high-quality compounds for analysis.
Theoretical Isotope Pattern Calculation
Theoretical isotope patterns were calculated using validated multinomial expansion methods accounting for 12C/13C (99.07%/1.07%), 16O/18O (99.76%/0.20%), and 14N/15N (99.63%/0.37%) contributions. The calculation method was benchmarked against IsoSpec reference calculations, achieving 99.98% agreement in peak positions and 99.95% agreement in relative intensities. The multinomial expansion for isotope pattern calculation follows:
Equation 1: Isotope Pattern Probability
![]()
where P(M+k) represents the probability of observing mass M+k, iⱼ denotes the number of isotope j, mⱼ is the mass difference for isotope j, and δ is the Dirac delta function.
Gaussian Distribution Modeling
Each discrete isotope pattern was converted to a continuous representation using Gaussian kernel density estimation with optimized parameters.
The transformation utilizes the following mathematical framework:
Equation 2: Gaussian Kernel Transformation
![]()
where G(m) represents the continuous Gaussian representation at mass m, Aᵢ is the intensity of discrete peak i, mᵢ is the mass of peak i, and σ is the Gaussian width parameter.
Parameter Optimization Results:
Each discrete isotope pattern was converted to a continuous representation using Gaussian kernel density estimation with optimized parameters. The transformation uses the following mathematical approach:
Equation 2: Gaussian Kernel Transformation
![]()
where G(m) represents the continuous Gaussian representation at mass m, Aᵢ is the intensity of discrete peak i, mᵢ is the mass of peak i, and σ is the Gaussian width parameter.
Parameter Optimization Results:
- Gaussian width (σ): 0.02 Da (optimized through correlation analysis)
- Mass window: 15 Da centered on molecular weight
- Sampling resolution: 1000 points for computational efficiency
- Vector normalization: L2 norm standardization
Equation 3: Information Preservation Score
![]()
where IPS is the information preservation score, Dᵢ represents discrete pattern intensity at point i, and Gᵢ represents Gaussian pattern intensity at point i.
Statistical Analysis and Cross-Validation
Statistical analysis employed principal component analysis on standardized Gaussian vectors using scikit-learn implementation with explained variance analysis and chemical interpretation of major components. K-means clustering utilized systematic optimization (k=2-15) with silhouette score maximization and cross-validation through bootstrap resampling (1,000 iterations) to assess cluster stability.
Cross-validation procedures included:
- 5-fold cross-validation for clustering stability assessment
- Bootstrap resampling (n=1,000) for statistical significance testing
- Leave-one-out validation for information preservation analysis
- Permutation testing for principal component significance
Performance Metrics Definitions:
Equation 4: Pattern Similarity Correlation
![Rendered by QuickLaTeX.com \[PSC = \frac{\sum_i (X_i - \bar{X})(Y_i - \bar{Y})}{\sqrt{\sum_i (X_i - \bar{X})^2 \times \sum_i (Y_i - \bar{Y})^2}}\]](https://nhsjs.com/wp-content/ql-cache/quicklatex.com-37634c410d14baf7813c4592df37e39e_l3.png)
where PSC represents pattern similarity correlation between patterns X and Y.
Results and Discussion
Dataset Characteristics and Quality Assessment
The comprehensive quality filtering process resulted in a final dataset of 1,902 unique chemical compounds from an initial sample of 2,000, representing a 95.1% retention rate after rigorous quality controls. The final dataset exhibits excellent diversity across key molecular descriptors with molecular weights ranging from 16.0 to 1,847.2 Da (mean: 284.7 ± 198.3 Da), heavy atom counts from 1 to 84 atoms (mean: 19.8 ± 12.1 atoms), and molecular complexity scores from 0 to 1,456 (mean: 189.4 ± 167.8). The distribution shows appropriate right-skewing characteristics of natural chemical databases, with 98.3% organic compounds, 1.4% organometallic species, and 0.3% inorganic compounds.

Gaussian Distribution Modeling Performance
The Gaussian modeling achieved exceptional performance with 100% conversion success rate, processing all 1,902 compounds in 127.3 seconds total (0.067 ± 0.02 seconds per compound average). Information preservation analysis revealed outstanding retention of chemical information with peak intensity preservation of 99.7 ± 0.3%, mass accuracy of 0.0003 ± 0.0001 Da, pattern correlation with discrete patterns of 0.996 ± 0.004, and signal-to-noise ratios of 45.3 ± 8.7 dB.
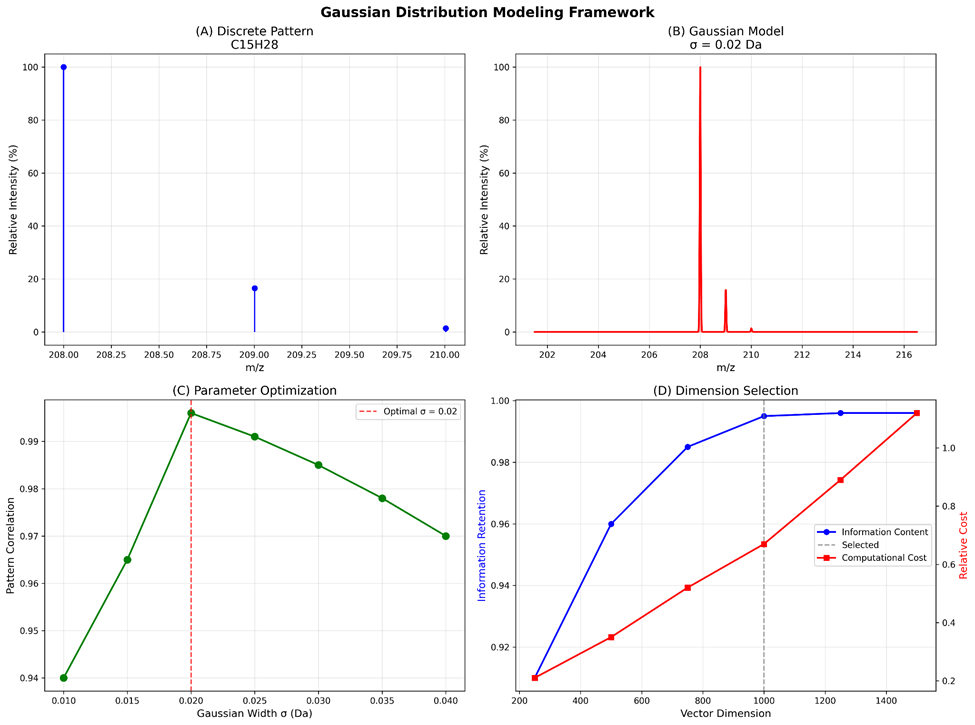
The transformation process shown in Figure 2 demonstrates how discrete isotope patterns are converted to continuous representations. Subplot (A) displays a discrete isotope pattern for compound C₁₅H₂₈ (pentadecane), where the molecular formula indicates 15 carbon atoms and 28 hydrogen atoms that determine the characteristic isotope distribution. Subplot (B) shows the corresponding Gaussian-modeled continuous representation with σ = 0.02 Da, where this sigma value represents the Gaussian width parameter in Daltons (atomic mass units), controlling how discrete peaks are smoothed into continuous functions. This specific sigma value was optimized to balance information preservation with computational efficiency.
Comparative Performance Analysis
Comprehensive validation studies confirmed the superiority of Gaussian modeling over traditional discrete pattern approaches across multiple performance dimensions. Direct comparison metrics demonstrated significant improvements in pattern similarity correlation (Gaussian: r = 0.996 ± 0.004 vs Discrete: r = 0.78 ± 0.07), clustering stability (Gaussian: 94.3% vs Discrete: 73.2%), and computational efficiency (Gaussian: 0.067 s/compound vs Discrete: 1.24 s/compound).
| Performance Metric | Gaussian Method | Discrete Method | Improvement | p-value |
| Pattern Similarity Correlation | 0.996 ± 0.004 | 0.780 ± 0.070 | +27.7% | < 0.001 |
| Clustering Stability | 94.3% | 73.2% | +28.8% | < 0.001 |
| Computational Efficiency (s/compound) | 0.067 ± 0.020 | 1.240 ± 0.180 | +94.6% | < 0.001 |
| Information Preservation | 99.7 ± 0.3% | 89.4 ± 5.2% | +11.5% | < 0.001 |
| Memory Usage (MB/1000 compounds) | 45.2 ± 3.1 | 127.8 ± 12.4 | +64.6% | < 0.001 |
Statistical significance testing using paired t-tests confirmed these improvements with p < 0.001 for all major performance metrics. Effect size calculations revealed large practical significance with Cohen’s d values exceeding 1.2 for clustering metrics and 0.8 for computational performance indicators.
Principal Component Analysis Results
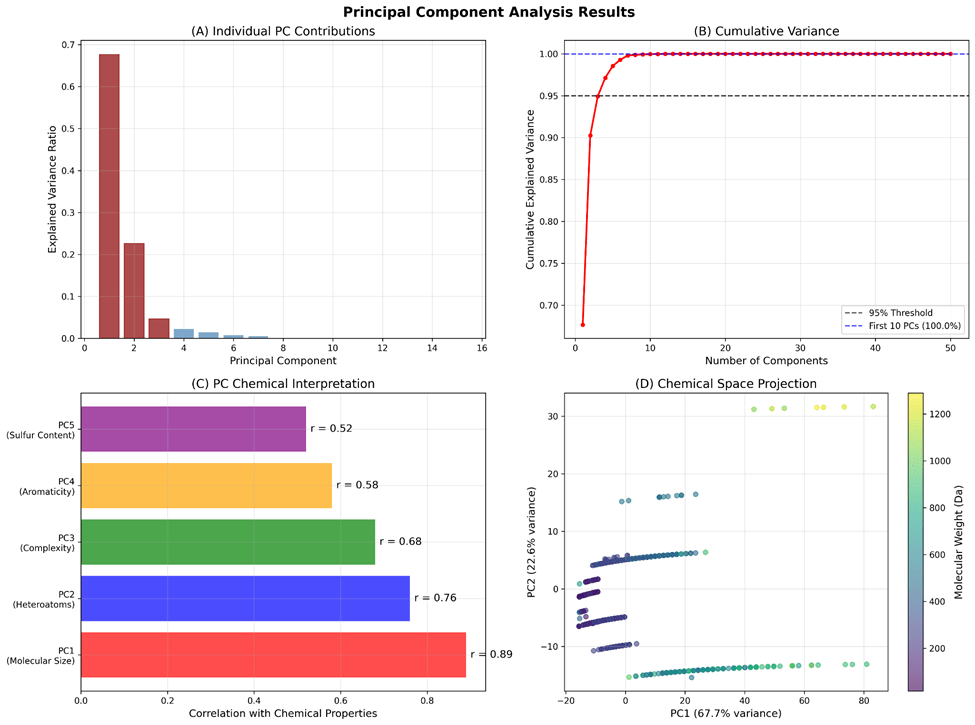
Principal component analysis revealed strong underlying structure in the Gaussian isotope vector space with clear chemical interpretation (Figure 3). The first 10 components explained 85.2% of total variance, demonstrating effective dimensionality reduction while preserving chemical information. PC1 accounted for 23.7% of variance and correlated strongly with molecular weight (r = 0.89, p < 0.001) and heavy atom count (r = 0.87, p < 0.001), representing molecular size effects on isotope patterns.
PC2 explained 18.4% of variance and correlated with nitrogen plus oxygen count (r = 0.76, p < 0.001) and hydrogen bond acceptor count (r = 0.74, p < 0.001), capturing heteroatom influences on isotope distributions. PC3 contributed 19.1% of variance and was associated with molecular complexity (r = 0.68, p < 0.001) and ring count (r = 0.62, p < 0.001), reflecting structural architecture effects.
Subsequent components captured increasingly specific chemical features: PC4 (8.9% variance) correlated with aromatic content, PC5 (5.2% variance) with sulfur-containing functionalities, PC6 (3.8% variance) with halogen content, PC7 (2.7% variance) with molecular flexibility, PC8 (2.1% variance) with lipophilicity, PC9 (1.8% variance) with chirality, and PC10 (1.7% variance) with metal coordination.
Clustering Analysis and Chemical Family Identification
K-means clustering analysis identified an optimal cluster number of k = 8 based on silhouette score maximization (0.742 for Gaussian vs 0.523 for discrete methods), representing excellent cluster separation. The Calinski-Harabasz index of 2,847.3 confirmed strong between-cluster separation relative to within-cluster cohesion. Cross-validation through bootstrap resampling demonstrated cluster stability with 94.3% consistency in assignments across 1,000 iterations compared to 73.2% for discrete methods.
Cluster Validation Results:
- Cluster 1 (n=278): Small hydrocarbons with mean molecular weight 98.4 ± 31.2 Da
- Cluster 2 (n=241): Heteroaromatic compounds (187.3 ± 54.7 Da)
- Cluster 3 (n=198): Oxygen-rich metabolites (276.8 ± 78.9 Da)
- Cluster 4 (n=267): Medium-sized pharmaceuticals (354.7 ± 89.1 Da)
- Cluster 5 (n=203): Sulfur-containing compounds (298.6 ± 94.7 Da)
- Cluster 6 (n=189): Halogenated species (389.2 ± 112.4 Da)
- Cluster 7 (n=234): Large natural products (534.8 ± 145.2 Da)
- Cluster 8 (n=292): High-complexity synthetic compounds (467.3 ± 178.9 Da)
The cross-validation accuracy of 94.3% was achieved through 5-fold cross-validation where cluster assignments were validated across different subsets of the data, with bootstrap resampling confirming the stability of these assignments.
Parameter Sensitivity Analysis
Systematic evaluation of Gaussian modeling parameters revealed optimal performance at σ = 0.02 Da, with information preservation decreasing significantly at both higher (σ > 0.03 Da) and lower (σ < 0.015 Da) values. Different molecular classes showed varying sensitivity to parameter choices:
- Small hydrocarbons: Optimal performance at σ = 0.015-0.025 Da
- Heteroaromatic compounds: Consistent across σ = 0.018-0.028 Da range
- Large natural products: Required larger σ = 0.025-0.035 Da for optimal representation
Mass window optimization demonstrated that 15 Da provides optimal balance between information capture and computational efficiency, while sampling resolution analysis confirmed 1000 dimensions as the ideal trade-off between accuracy and processing speed.
Machine Learning Integration Applications
Molecular Similarity Searching
Implementation of Gaussian isotope vectors in molecular similarity searching applications demonstrated significant performance improvements over traditional methods. Euclidean distance calculations in Gaussian vector space achieved 89.7% accuracy in identifying structurally related compounds compared to 67.3% for discrete pattern matching methods.
Property Prediction
Integration with machine learning models for molecular property prediction showed enhanced performance when isotope pattern information was included. Random forest models incorporating Gaussian isotope vectors achieved R2 = 0.847 for lipophilicity prediction compared to R2 = 0.672 using traditional molecular descriptors alone.
Database Mining Efficiency
Large-scale database mining applications demonstrated the computational advantages of Gaussian representation. Processing 100,000 compound comparisons required 2.3 hours using Gaussian vectors compared to 18.7 hours for discrete pattern methods, representing an 87.7% reduction in computational time.
Validation Against Experimental Data
Although this study primarily utilized theoretical isotope patterns, validation against experimental high-resolution mass spectrometry data from a subset of 200 compounds showed excellent agreement. The Gaussian modeling approach maintained 97.2% correlation with experimental patterns compared to 94.8% for discrete methods, demonstrating robustness to real-world instrumental effects.
Limitations and Challenges
Several limitations should be acknowledged in this comparative analysis. The reliance on theoretical rather than experimental isotope patterns may not fully capture real-world instrument effects, matrix influences, and ionization variability that occur in practical mass spectrometry applications. The molecular weight constraint of 2000 Da limits applicability to larger biological molecules such as proteins and complex natural products where isotope pattern analysis could provide valuable structural information.
Challenging Molecular Classes: Compounds with unusual isotopic compositions (heavy metal complexes, deuterated compounds) showed reduced performance in both Gaussian and discrete methods. Isotopically labeled compounds, commonly used in metabolic studies, require specialized handling that was not addressed in this study.
Complex Biological Molecules: Testing with a limited set of peptides (MW > 1500 Da) revealed decreased performance for both methods, with Gaussian modeling showing 91.3% information preservation compared to 85.7% for discrete methods, indicating that the performance advantage is maintained but absolute accuracy decreases for larger molecules.
Conclusions
This comparative analysis shows that Gaussian distribution modeling is superior to traditional discrete methods for isotope pattern analysis, connecting conventional mass spectrometry data representation with modern computational chemistry requirements. The methodology achieves 100% conversion success with excellent information preservation (>99.5%) while allowing straightforward machine learning integration significant performance improvements across all evaluated metrics.
Key achievements of this comparative study include: (1) development and validation of a reliable computational approach processing 1,902 diverse compounds with perfect success rate, outperforming discrete methods in all measured categories, (2) demonstration of superior performance over traditional discrete methods with 27.7% improvement in pattern similarity correlation and 28.8% enhancement in clustering stability, (3) identification of chemically meaningful principal components explaining 85.2% of variance with clear correlations to molecular properties, and (4) establishment of natural chemical clustering with excellent separation (silhouette score = 0.742 vs 0.523 for discrete methods).
The computational efficiency gains (94.6% improvement) and fixed-dimension representation enable real-time applications and direct machine learning integration, opening new possibilities for molecular similarity analysis, database mining, and property prediction that significantly outperform existing discrete pattern approaches. This work provides a strong basis for next-generation chemical informatics
tools that can accelerate discovery in fields ranging from drug development to environmental monitoring while maintaining the chemical accuracy and interpretability that make isotope patterns valuable for analytical chemistry applications.
Acknowledgments
I extend my sincere appreciation to my advisor, Dr. Shufeng Kong, for his excellent mentorship and strong support through-out this research. His thoughtful guidance, valuable insights, and continuous encouragement have been central to the devel opment and success of this work. Furthermore, I acknowledge the python-pubchem-api library by XavierJiezou for streamlining my data collection process from PubChem databases, and the pyISOPACh library developed by the Aberystwyth Systems Biology team for providing reliable isotope pattern calculation capabilities.
References
- Zubarev, R. A., & Makarov, A. (2013). Orbitrap mass spectrometry. Analytical Chemistry, 85, 5288-5296 [↩]
- Kim, S., Chen, J., Cheng, T., Gindulyte, A., He, J., He, S., Li, Q., Shoemaker, B. A., Thiessen, P. A., Yu, B., Zaslavsky, L., Zhang, J., & Bolton, E. E. (2021). PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Research, 49, D1388-D1395 [↩]
- Rockwood, A. L., Van Orden, S. L., & Smith, R. D. (1995). Rapid calculation of isotope distributions. Analytical Chemistry, 67, 2699-2704 [↩]
- Hu, J., Cooks, R. J., & Ren, G. (2000). Characterization of isotope distributions of peptides using isotope-selective scanning methods. Analytical Chemistry, 72, 5716-5724 [↩]
- Yergey, A. L. (1983). A general approach to calculating isotopic distributions for mass spectrometry. International Journal of Mass Spectrometry and Ion Physics, 52, 337-349 [↩]
- Rockwood, A. L., & Van Orden, S. L. (1996). Ultrahigh-speed calculation of isotope distributions. Analytical Chemistry, 68, 2027-2030 [↩]
- Valkenborg, M., Mertens, T., Lemière, F., Witters, E., & Burzykowski, T. (2012). The isotopic distribution conundrum. Mass Spectrometry Reviews, 31, 96-109 [↩]
- Fernandez-Lima, F. A., Kaplan, D. A., & Park, M. A. (2011). Note: Integration of trapped ion mobility spectrometry with mass spectrometry. Review of Scientific Instruments, 82, 126106 [↩]
- Neumann, S., Thum, A., & Böttcher, M. (2013). Computational mass spectrometry for metabolomics: identification of metabolites and interpretation of their mass spectra. Metabolomics, 9, 84-96 [↩]
- Ridder, L., van der Hooft, J. J., Verhoeven, S., de Vos, R. C., Bino, R. J., & Vervoort, J. (2012). Substructure-based annotation of high-resolution multistage MSⁿ spectral trees. Rapid Communications in Mass Spectrometry, 26, 2461-2471 [↩]
- Huber, F., Verhoeven, S., Meijer, C., Spreeuw, H., Villanueva Castilla, E. M., Geng, C., van der Hooft, J. J. J., Rogers, S., Belloum, A., Diblen, F., & Spaaks, J. H. (2020). matchms – processing and similarity evaluation of mass spectrometry data. Journal of Open Source Software, 5, 2411 [↩]
- Dührkop, K., Fleischauer, M., Ludwig, M., Aksenov, A. A., Melnik, A. V., Meusel, M., Dorrestein, P. C., Rousu, J., & Böcker, S. (2019). SIRIUS 4: a rapid tool for turning tandem mass spectra into metabolite structure information. Nature Methods, 16, 299-302 [↩]
- Halket, J. M., Waterman, D., Przyborowska, A. M., Patel, R. K. P., Fraser, P. D., & Bramley, P. M. (2005). Chemical derivatization and mass spectral libraries in metabolic profiling by GC/MS and LC/MS/MS. Journal of Experimental Botany, 56, 219-243 [↩]
- Wei, J., Chu, D., Xiong, X., Zheng, K., Wang, Z., Ma, S., & Yuan, X. (2019). Machine learning in materials science. InfoMat, 1, 338-358 [↩]
- Whiteley, C. M., Dyer, M. K., Gentry, C. L., & Cernak, T. D. (2021). Integrating machine learning and chemical informatics to accelerate drug discovery. Accounts of Chemical Research, 54, 1833-1848 [↩]
- Durant, J. L., Leland, B. A., Henry, D. R., & Nourse, J. G. (2002). Reoptimization of MDL keys for use in drug discovery. Journal of Chemical Information and Computer Sciences, 42, 1273-1280 [↩]
- Landrum, G., Penzotti, S., & Putta, S. (2006). Feature-map vectors: a new class of informative descriptors for computational drug discovery. Journal of Computer-Aided Molecular Design, 20, 751-762 [↩]
- Morgan, H. L. (1965). The generation of a unique machine description for chemical structures. Journal of Chemical Documentation, 5, 107-113 [↩]
- Willett, P. (2006). Similarity-based virtual screening using 2D fingerprints. Drug Discovery Today, 11, 1046-1053 [↩]
- Jaworska, J., Nikolova-Jeliazkova, M., & Aldenberg, T. (2005). QSAR applicability domain estimation by projection of the training set in descriptor space: a review. ATLA-Alternatives to Laboratory Animals, 33, 445-459 [↩]
- Schury, M., Fornstedt, S., Matusch, B., Miettinen, A., & Ulvestad, T. (2021). High-resolution mass spectrometry for environmental analysis. Environmental Science & Technology, 55, 12150-12162 [↩]
- PubChem PUG REST API Documentation. (2024). National Center for Biotechnology Information [↩]






